Exámenes genéticos para la prevención de la erupción cutánea grave inducida por fármacos
Resumen
Antecedentes
Las reacciones cutáneas inducidas por fármacos se presentan con una serie de síntomas clínicos, desde erupciones cutáneas maculopapulares leves hasta erupciones cutáneas ampollares graves, como el síndrome de Stevens‐Johnson (SSJ) o la necrólisis epidérmica tóxica (NET), que pueden provocar la muerte. Las reacciones más leves pueden ser problemáticas y llevar a un bajo nivel de adherencia a los medicamentos. La patogénesis de estas reacciones a los medicamentos aún no se comprende del todo; sin embargo, hay evidencia de que los exámenes genéticos previos al tratamiento pueden ayudar a predecir y prevenir estas reacciones en algunos casos.
Objetivos
Evaluar los efectos del cribado farmacogenético prospectivo para reducir las reacciones cutáneas asociadas al fármaco en una población de pacientes.
Métodos de búsqueda
Se hicieron búsquedas en las siguientes bases de datos hasta julio 2018: Registro Especializado Cochrane de Piel, CENTRAL, MEDLINE, Embase y LILACS. También se buscó en cinco registros de ensayos, y se verificaron las listas de referencias de los estudios incluidos y revisiones relevantes para obtener más referencias de ensayos controlados aleatorios (ECA).
Criterios de selección
Se incluyeron ECA de participantes sometidos a cribado farmacogenético prospectivo para determinar variantes genéticas asociadas con reacciones de hipersensibilidad, en comparación con los que no se sometieron a cribado farmacogenético prospectivo. Se incluyeron participantes de cualquier ámbito, de cualquier edad, sexo y origen étnico, que habían recibido una prescripción de fármacos que se sabe provocan reacciones de hipersensibilidad de tipo retardado.
Obtención y análisis de los datos
Se utilizaron los procedimientos metodológicos estándar previstos por Cochrane. Para evaluar la inclusión de los estudios, dos autores de revisión cribaron de forma independiente todos los títulos y resúmenes de las publicaciones identificadas mediante las búsquedas. Debido a que solo hubo un estudio incluido, muchos de los análisis de datos planificados no fueron aplicables a la revisión. Se utilizaron los criterios GRADE para evaluar la calidad del estudio incluido.
Los resultados primarios de la revisión fueron la incidencia de erupciones cutáneas graves con síntomas sistémicos (como fiebre y compromiso de múltiples órganos) y efectos a largo plazo (como cicatrización de los párpados o del tejido pulmonar). Los resultados secundarios fueron la hospitalización para las reacciones cutáneas inducidas por fármacos, las reacciones cutáneas ampollares (como el SSJ, el síndrome de hipersensibilidad [HSS]) y la muerte.
Resultados principales
Un estudio (un ensayo aleatorizado, doble ciego, controlado y multicéntrico) cumplió con los criterios de inclusión. El ensayo incluyó a 1956 participantes adultos (74% hombres, con una media de edad de 42 años) de 265 centros (centros médicos, hospitales, clínicas ambulatorias) en 19 países de todo el mundo, con infección por el VIH tipo 1 y que no habían recibido abacavir anteriormente. Los participantes, que tenían una necesidad clínica de tratamiento con un régimen de medicamentos antirretrovirales que incluía el abacavir, fueron asignados al azar para someterse a un cribado del antígeno leucocitario humano prospectivo (HLA) Clase I, locus B, alelo 57:01 (HLA‐B*57:01) (grupo de detección prospectiva) antes de este tratamiento, o para someterse a un enfoque de atención estándar de uso de abacavir sin un cribado prospectivo del HLA‐B*57:01 (grupo de control). A los participantes con resultados positivos del examen de HLA‐B*57:01 no se les administró abacavir; en cambio, recibieron terapia antirretroviral sin este fármaco. Al grupo de control se le realizó un examen farmacogenético retrospectivo de HLA‐B*57:01. La duración del ensayo fue de seis meses. Cada participante fue observado durante seis semanas. Las evaluaciones se realizaron en el momento del ingreso al estudio, al inicio (día uno del tratamiento con abacavir) y en las semanas uno, dos y seis. Este estudio fue financiado por el fabricante de abacavir, GlaxoSmithKline.
El estudio no evaluó ninguno de los resultados primarios y no midió ninguno de los resultados secundarios de forma aislada. Sin embargo, sí evaluó un resultado de reacción de hipersensibilidad (característicamente grave) que incluyó (pero no se limitó a) los resultados secundarios de HSS y SSJ/NET.
El estudio demostró que el cribado prospectivo de HLA‐B*57:01 probablemente reduce la incidencia de la reacción de hipersensibilidad al abacavir. La incidencia de la reacción de HSS clínicamente diagnosticada al abacavir fue menor en el brazo de cribado (cociente de riesgos [CR] 0,43; intervalo de confianza [IC] del 95%: 0,28 a 0,67; 1650 participantes; evidencia de calidad moderada), al igual que la reacción de HSS confirmada inmunológicamente (CR 0,02; IC del 95%: 0,00 a 0,37; 1644 participantes; evidencia de calidad moderada). El resultado positivo de una prueba de parche epicutánea realizada de seis a diez semanas después del diagnóstico clínico fue la confirmación inmunológica.
En general, el estudio demuestra un riesgo de sesgo bajo en cinco de los siete dominios. Hubo un riesgo de sesgo de detección alto porque las reacciones de hipersensibilidad fueron diagnosticadas por el investigador principal en el sitio de reclutamiento sin el uso de criterios clínicos predefinidos. Aunque también hubo un riesgo de sesgo de desgaste alto debido a la exclusión de los participantes con seguimiento incompleto de los análisis, los autores realizaron una serie de análisis de sensibilidad basados en la población con intención de tratar, que demostraron resultados congruentes con el análisis primario. Se calificó la calidad del estudio como moderada mediante los criterios GRADE.
Conclusiones de los autores
El cribado prospectivo de HLA‐B*57:01 probablemente reduce las reacciones cutáneas de hipersensibilidad severas al abacavir en pacientes con pruebas positivas para el VIH tipo 1. Sin embargo, estos resultados solo se basan en un estudio, que tuvo un riesgo de abandono y de detección alto.
Los resultados primarios (incidencia de erupciones cutáneas graves con síntomas sistémicos y efectos a largo plazo) no fueron evaluados por el ensayo, y solo se midió uno de los resultados secundarios de la revisión (reacción de hipersensibilidad); por lo tanto, no se encontró evidencia relacionada con la hospitalización, la muerte ni las afecciones a largo plazo como resultado de la lesión por fármacos.
No se encontró evidencia elegible sobre los exámenes genéticos para la erupción cutánea grave inducida por fármacos en relación con diferentes fármacos y clases de fármacos. Los ensayos clínicos adicionales basados en otros fármacos, y en diferentes poblaciones de pacientes, serían útiles para aconsejar cambios de política con objeto de mejorar la prevención de las reacciones cutáneas adversas a los tratamientos farmacológicos.
PICO
Resumen en términos sencillos
Exámenes genéticos para predecir y prevenir las erupciones cutáneas graves causadas por fármacos
Pregunta de la revisión
El objetivo de esta revisión Cochrane es determinar si los exámenes genéticos para ciertos genotipos (es decir, la presencia o ausencia de una variación específica en un gen) antes de indicar los fármacos pueden prevenir reacciones cutáneas graves en pacientes que toman fármacos que se sabe causan reacciones de hipersensibilidad tipo retardada (es decir, erupción cutánea alérgica hasta seis semanas después de tomar la medicación prescrita). Se comparó a los pacientes sometidos a este tipo de examen con los pacientes en quienes no se realizó. Se buscaron todos los estudios relevantes, por lo que se los pudo analizar para responder esta pregunta. Sin embargo, solo se halló uno.
Antecedentes
Distintos tipos de medicamentos pueden causar efectos indeseables, incluidas las erupciones cutáneas. Por lo general, estas reacciones cutáneas son leves; sin embargo, en raras (pero posibles) ocasiones el fármaco puede causar desprendimiento de la piel, fiebre y compromiso de órganos internos, y estos efectos pueden ser mortales. Los casos graves pueden requerir hospitalización y tratamiento en unidades especializadas en quemaduras. No se comprende del todo cómo se producen estas reacciones y qué pacientes presentan un mayor riesgo, aunque se sabe que cumplen una función los factores genéticos. Se han llevado a cabo estudios de investigación sobre el uso de exámenes genéticos simples para predecir y prevenir estas reacciones.
Características de los estudios
Sólo se encontró un estudio relevante. Este estudio incorporó a 1956 participantes adultos, de los que el 74% eran hombres, con pruebas positivas para VIH tipo 1 y candidatos para iniciar una terapia antirretroviral altamente activa con un medicamento llamado abacavir. (Los antirretrovirales son una clase de medicamentos que se utilizan para tratar a los pacientes con infección por VIH.) La edad de los participantes del estudio varió de 18 a 77 años; la edad promedio fue de 42 años. Los participantes procedían de 19 países de todo el mundo en 265 centros sanitarios (por ejemplo, hospitales, clínicas). Este estudio investigó si los exámenes para el marcador genético HLA‐B*57:01 antes de prescribir abacavir (es decir, exámenes prospectivos) versus ningún examen genético prospectivo para HLA‐B*57:01 pueden reducir la tasa de reacciones cutáneas graves al fármaco. Se sabe que el HLA‐B*57:01 es clave para desarrollar reacciones cutáneas severas al medicamento antirretroviral contra el VIH, abacavir. A los participantes con exámenes positivos para el genotipo no se les administró abacavir, sino una terapia antirretroviral diferente. El estudio duró seis meses y cada participante fue observado durante seis semanas. GlaxoSmithKline, la compañía que fabrica abacavir, financió el estudio. Los participantes del grupo de control fueron sometidos al examen farmacogenético HLA‐B*5701 después de haber recibido abacavir como estándar de atención.
Resultados clave
El único estudio incluido no informó de datos para todos los participantes, y la evaluación clínica de la hipersensibilidad (HSS) se realizó en el momento del ingreso al estudio, al inicio (día uno) y a las semanas uno, dos y seis, sin utilizar criterios predefinidos. Estos pueden ser fuentes importantes de sesgo y, por lo tanto, se consideró que la calidad de la evidencia fue moderada. Los datos disponibles mostraron que el cribado prospectivo de HLA‐B*57:01 probablemente reduce la incidencia de la reacción de hipersensibilidad al abacavir (incluidos, aunque no limitados a, los resultados secundarios del síndrome de hipersensibilidad [HSS] y el síndrome de Stevens‐Johnson [SSJ] o la necrólisis epidérmica tóxica [NET] [es decir, reacciones cutáneas ampollares severas causadas por medicamentos]).
A partir de estos resultados, se esperaría que de 1000 individuos no sometidos a un cribado farmacogenético prospectivo (es decir, un cribado para evaluar la respuesta posible a un fármaco en particular), 78 presentarán una reacción de hipersensibilidad incluidos SSJ/NET (evaluación clínica), en comparación con 22 a 52 individuos sometidos a un cribado farmacogenético prospectivo con el examen de HLA‐B*57:01.
Además, se esperaría que de 1000 individuos no sometidos a un cribado farmacogenético prospectivo (atención estándar), 27 presentarán una reacción de hipersensibilidad incluido SSJ/NET (confirmado inmunológicamente), en comparación con ningún participante sometido a un cribado farmacogenético prospectivo con examen de HLA‐B*57:01. La prueba del parche cutáneo brindó confirmación inmunológica seis a diez semanas después del diagnóstico clínico.
El estudio no midió los otros resultados de esta revisión.
La evidencia está actualizada hasta julio 2018.
Calidad de las pruebas
El estudio incluido se calificó como de calidad moderada. Algunos pacientes fueron retirados del estudio y no incluidos en los análisis, y el investigador que diagnosticó las reacciones de HSS no utilizó un criterio clínico predefinido de hipersensibilidad. Estos problemas resultaron en una disminución de la calificación de la certeza de la evidencia.
Authors' conclusions
Summary of findings
| Prospective genetic HLA‐B*57:01 screening compared with standard care for drug‐induced skin rash | ||||||
| Patient or population: patients with HIV‐1 infection and a pre‐established clinical need for treatment with an antiretroviral drug regimen containing abacavir but with an unknown HLA‐B*57:01 status. Settings: secondary care clinics Intervention: prospective genetic screening for the HLA‐B*57:01 allele Comparison: no prospective genotyping, standard‐of‐care treatment | ||||||
| Outcomes | Illustrative comparative risks* (95% CI) | Relative effect | No of Participants | Quality of the evidence | Comments | |
| Assumed risk | Corresponding risk | |||||
| Standard Carea | Prospective genetic HLA‐B*57:01 screening | |||||
| Severe skin drug‐induced rash | No data | ‐ | ‐ | ‐ | Not assessed | |
| Long‐term sequelae | No data | ‐ | ‐ | ‐ | Not assessed | |
| Hospitalisation for drug‐induced skin reaction | No data | ‐ | ‐ | ‐ | Not assessed | |
| SJS/TEN (Stevens‐Johnson syndrome/toxic epidermal necrolysis) | See hypersensitivity | ‐ | ‐ | ‐ | Not assessable | |
| AGEP (acute generalised exanthematous pustulosis) | No data | ‐ | ‐ | ‐ | Not assessed | |
| HSS (hypersensitivity) reaction including SJS/TEN (clinically diagnosed) (6 weeks clinical assessment) | 78 per 1000 | 34 per 1000 | RR 0.43 (0.28 to 0.67) | 1650 participants | ⊕⊕⊕⊝ | ‐ |
| HSS reaction including SJS/TEN (immunologically confirmed) (6 weeks clinical assessment) | 27 per 1000 | 0 per 1000 | RR 0.02 (0.00 to 0.37) | 1644 participants (1 study) | ⊕⊕⊕⊝ moderateb | ‐ |
| Death | No data | ‐ | ‐ | ‐ | Not assessed | |
| *The basis for the assumed risk (e.g. the median control group risk across studies) is provided in footnotes. The corresponding risk (and its 95% confidence interval) is based on the assumed risk in the comparison group and the relative effect of the intervention (and its 95% CI). | ||||||
| GRADE Working Group grades of evidence | ||||||
| a The assumed risk is estimated from the event rate (confirmed clinically diagnosed occurrences of HSS including SJS/TEN or immunologically confirmed) in the control arm of the included study. b We downgraded the evidence to moderate quality, due to study limitations (high risk of detection and attrition bias). | ||||||
Background
We have explained some terms we have used in a glossary. Please see Table 1.
| Term | Explanation |
| Allele | One of two or more alternative forms of a gene at corresponding sites (loci) on homologous chromosomes |
| Antiretroviral | A class of drugs that inhibit the activity of retroviruses that cause HIV infection |
| Hardy‐Weinberg equilibrium | This states that allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences |
| HLA | Human leukocyte antigen: a group of protein molecules located on bone marrow and other cells that can provoke an immune response |
| Hypersensitivity | A state of altered reactivity in which the body reacts with an exaggerated immune response to a foreign substance, such as a drug |
| Immunologically confirmed | Patch testing is done to see whether a particular drug is causing allergic skin reaction. Patch test can detect delayed allergic or immunological reaction and confirm the diagnosis of hypersensitivity. |
| Phenotypes | The set of observable characteristics of an individual resulting from the interaction of its genotype with the environment |
| Polymorphic | A variation in the DNA that is too common to be due merely to new mutation. A polymorphism must have a frequency of at least 1% in a population |
| Maculopapular rash | A rash with both macules (flat and coloured like a freckle) and papules (a small raised spot) |
| Sequelae | A condition that is a consequence of a previous disease or injury |
| T‐cells | Another term for T‐lymphocyte, a type of cell that participates in immune response |
Description of the condition
Some drugs may cause skin rashes that vary in their severity and incidence. Skin reactions caused by drugs, often termed 'drug‐induced skin injury', are common (carbamazepine‐induced skin rash has a 10% incidence rate (Marson 2007)); they can present with a range of clinical manifestations, from a mild maculopapular skin rash to life‐threatening skin rashes such as Stevens‐Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) (Pirmohamed 2004; Roujeau 1987). The most severe forms of hypersensitivity are very rare, but these may result in up to 30% mortality (Roujeau 1994). Less severe forms of hypersensitivity reactions are still troublesome and may prevent people from taking medications that are otherwise effective. Delayed type hypersensitivity reactions are T‐cell mediated: they usually occur 24 to 48 hours, and up to six weeks, after exposure to culprit drugs.
The mechanisms involved in the pathogenesis of these drug‐induced reactions are still poorly understood; however, immunogenetic and non‐immune factors have been implicated (Chung 2004; Pirmohamed 2004; Vitezica 2008; Watanabe 2010). Recent evidence suggests that drug‐specific T‐cells can be identified in individuals who previously experienced adverse drug reactions to the culprit drug (Illing 2012).
Description of the intervention
There is increasing evidence from clinical trials that pretreatment genetic testing may reduce the possibility of severe drug‐induced hypersensitivity (Chen 2011; Mallal 2008).
Figure 1 represents a diagram of decision‐making informed by genetic testing.
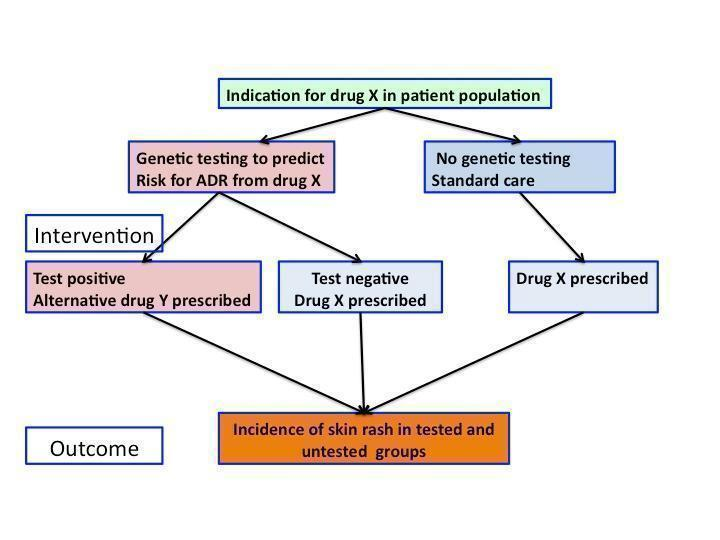
Flowchart of interventions (genetic testing) and outcomes (skin rash) in a patient population prescribed drug X
To date, the strongest association with drug‐induced skin injury has been reported with genetic variants in the human leukocyte antigens (HLA) (Amstutz 2013; Chung 2004; Hetherington 2002; Hung 2005; Mallal 2002; McCormack 2011; Ozkaya‐Bayazit 2001). HLA are cell surface proteins involved in presenting antigens to the immune system. They are encoded by most polymorphic genes in the human genome. However, different genetic markers are associated with hypersensitivity in different populations, and the effect size varies in different ethnicities. Also, there is evidence that some genetic factors could predispose to drug‐induced skin injury irrespective of the underlying drug aetiology. In addition, it is possible that different severity phenotypes can share the same predisposing factor (McCormack 2011). Table 2 shows reported associations between hypersensitivity reactions, which include skin injury and genetic variants in HLA genes.
| Drugs associated with skin injury | Class of drug | HLA allele | Population | Reference |
| Stevens‐Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) | ||||
| Allopurinol | Antiuric acid | B*5801 | Han Chinese | |
| ‐ | ‐ | ‐ | Thai | |
| ‐ | ‐ | ‐ | Japanese | |
| ‐ | ‐ | ‐ | Malay | |
| Carbamazepine | Antiepileptic | B*1502 | Han Chinese | |
| ‐ | ‐ | ‐ | Thai | Kulkantrakorn 2012; Locharernkul 2008; Tassaneeyakul 2010; Tangamornsuksan 2013 |
| ‐ | ‐ | ‐ | Malay | |
| ‐ | ‐ | ‐ | Indian | |
| ‐ | ‐ | A*3101 | White | |
| ‐ | ‐ | A*3101 | Japanese | |
| Phenytoin | Antiepileptic | B*1502 | Han Chinese | |
| ‐ | ‐ | ‐ | Thai | |
| Oxicam | Non‐steroidal anti‐inflammatory drug (NSAID) | A2, B12 | White | |
| Sulphamethoxazole | Antibiotic | A29, B12, DR7 | White | |
| Hypersensitivity syndrome (drug‐induced hypersensitivity syndrome (DIHS) or drug reaction with eosinophilia and systemic symptoms (DRESS)) | ||||
| Abacavir | Antiretroviral | B*57:01 | White | Hetherington 2002; Hughes 2004a; Mallal 2002; Mallal 2008; Martin 2004 |
| ‐ | ‐ | ‐ | African American | |
| Aminopenicillins | Antibiotic | A2, Drw52 | White | |
| Nevirapine | Antiretroviral | DRB1*01 | White ‐ Australian | |
| ‐ | ‐ | DRB1*01 | White ‐ French | |
| ‐ | ‐ | Cw8, B14 | White ‐ Italian | |
| ‐ | ‐ | Cw8 | Japanese | |
| ‐ | ‐ | B*3505 | Thai | |
| ‐ | ‐ | Cw4 | Thai | |
| ‐ | ‐ | C*0404 | Black African | |
| ‐ | ‐ | Cw*04 | Chinese | |
| Aspirin | NSAIDs | DRB1*1302, DQB1*0609 | ‐ | |
| ‐ | NSAIDs | DR11 | ‐ | |
| Iodine contrast media | ‐ | DR | White ‐ Spanish | |
| Paraphenylenediamine | Hair dye | DP | White ‐ German | |
| Gold sodium thiomalate | Treatment of rheumatoid arthritis | DR5 | White ‐ Spanish | |
| Lamotrigine | Antiepileptic | B*5801, A*6801 | White | |
| Trichloroethylene | Industrial solvent, dry cleaning | B*1301 | Japanese | |
| Fixed drug eruptions | ||||
| Co‐trimoxazole | Antibiotic | A30, B13, Cw6 | White ‐ Turkish | |
| Feprazone | Analgesic | B22 | ‐ | |
Standard clinical practice does not include genetic testing for most drugs, despite some strong evidence on the benefits of pretreatment genotyping (Kim 2005; McCormack 2011; Ozeki 2011). There are potential ethical issues with genetic tests, such as the effect on family members, unintended findings, and storage and access of data.
How the intervention might work
Two recent clinical trials suggested that pretreatment genetic testing could reduce the possibility of severe hypersensitivity induced with an anti‐AIDS drug, abacavir (Mallal 2008), and an antiepileptic drug, carbamazepine (Chen 2011).
Patients who have a clinical requirement for a particular drug treatment can be stratified on the basis of a genetic test. Those who test positive for the risk marker are not prescribed the culprit drug, while those who test negative are safe to take the medicine of interest. In this way, it may be possible to reduce the incidence of severe drug skin reactions in the genotyped group compared to the randomly assigned group of patients who are not offered genetic testing, but for whom decision on drug choice is based on traditional clinical and biochemical parameters (Chen 2011; Mallal 2008).
Why it is important to do this review
Adverse drug reactions affecting the skin are common; they can have high morbidity and mortality and are a burden for healthcare systems around the world. If we were able to predict these reactions using a simple genetic test, it should be possible to prevent them with one of the following approaches:
-
by prescribing alternative therapy, if available;
-
by informing patients and healthcare providers so the patients could be monitored more closely if they are at an increased risk;
-
by informing drug developers, in order to improve drug design and future drug development.
We aimed to assess current research evidence to determine whether prospective pharmacogenetic screening is effective in reducing drug‐associated skin reactions. The planned methods for this review were published as a protocol (Alfirevic 2014).
Objectives
To assess the effects of prospective pharmacogenetic screening to reduce drug‐associated skin reactions in a patient population.
Methods
Criteria for considering studies for this review
Types of studies
We included randomised controlled trials (RCTs) only.
Types of participants
We included participants who were prescribed drugs known to cause delayed type hypersensitivity reactions with skin involvement. We accepted participants in any setting, who were of any age, gender, and ethnicity.
Prescribed drugs included, but were not limited to: antiepileptic drugs, antiretroviral drugs, antigout drugs, and antibiotics such as beta‐lactams (penicillin, amoxicillin, piperacillin, cephalosporins) and sulphonamides (sulphamethoxazole and trimethoprim). We would have included studies that described only a subset of relevant participants. Decisions to include studies that only partially addressed the population of interest would have been documented in the review, and we would have conducted a sensitivity analysis to assess the impact of the decisions on the review's findings.
Types of interventions
We considered genetic testing for any genetic variants associated with hypersensitivity reactions using all available techniques to determine individual genotypes, and compared to standard clinical practice, which does not include a genetic test. The intervention was a randomly allocated genetic test; if the test was positive, a drug that can cause hypersensitivity was avoided. We included studies whose purpose was to genotype on the basis of likely adverse skin reaction. We excluded any participants who had previously been administered the study drugs.
Types of outcome measures
We based core outcome measures on several papers describing clinical classification of drug‐induced skin reactions, including a paper entitled 'Phenotype standardisation for immune‐mediated drug‐induced skin injury' (Pirmohamed 2011), as well as papers by the RegiSCAR (European Registry of Severe Cutaneous Adverse Reactions) consortium (Bouvresse 2012; Kardaun 2013; Sekula 2011).
We assessed clinically defined hypersensitivity reaction, immunologically confirmed hypersensitivity reactions (if skin patch testing or lymphocyte proliferation assay data were available), and long‐term sequelae (including ophthalmologic, cutaneous, or liver damage, etc.).
We have provided a full list of clinically relevant outcomes, and distinction between primary and secondary outcomes, in Appendix 1.
Primary outcomes
-
The incidence of severe drug‐induced skin rash (defined as skin rash with systemic symptoms including fever and multiple organ involvement).
-
Long‐term sequelae (any of the following: ophthalmologic, cutaneous, or liver damage, etc.) up to 12 months after the severe drug‐induced skin rash.
Secondary outcomes
-
Hospitalisation for drug‐induced skin reaction within three months of exposure to the drug.
-
SJS/TEN (Stevens‐Johnson syndrome, toxic epidermal necrolysis).
-
AGEP (acute generalised exanthematous pustulosis).
-
HSS (hypersensitivity syndrome).
-
Death.
Additional terminology for HSS includes the following: drug‐induced hypersensitivity syndrome (DIHS), drug reaction with eosinophilia and systemic symptoms (DRESS), drug‐induced delayed multiorgan hypersensitivity syndrome, and hypersensitivity reaction.
Search methods for identification of studies
We aimed to identify all relevant RCTs regardless of language or publication status (published, unpublished, in press, or in progress).
Electronic searches
The Cochrane Skin Information Specialist searched the following databases up to 9 July 2018 using strategies based on the draft strategy for MEDLINE in our published protocol (Alfirevic 2014):
-
the Cochrane Skin Specialised Register using the search strategy in Appendix 2;
-
the Cochrane Central Register of Controlled Trials (CENTRAL) 2018, Issue 6, in the Cochrane Library using the strategy in Appendix 3;
-
MEDLINE via Ovid (from 1946) using the strategy in Appendix 4;
-
Embase via Ovid (from 1974) using the strategy in Appendix 5; and
-
LILACS (Latin American and Caribbean Health Science Information database, from 1982) using the strategy in Appendix 6.
Trials registers
We (AA and AJ) searched the following trials registers on 15 July 2018 using the following terms: skin rash, genetic test, drug:
-
the ISRCTN registry (www.isrctn.com);
-
ClinicalTrials.gov (www.clinicaltrials.gov);
-
the Australian New Zealand Clinical Trials Registry (www.anzctr.org.au);
-
the World Health Organization International Clinical Trials Registry platform (www.who.int/trialsearch); and
-
the EU Clinical Trials Register (www.clinicaltrialsregister.eu/).
Searching other resources
References from included studies
We checked the bibliographies of the included studies and published reviews for further references to relevant trials.
Correspondence
We requested relevant information from one author of an included study, but they did not reply to us (Appendix 7).
Adverse effects
We did not perform a separate search for adverse effects of the target intervention. We examined data on adverse effects of genotyping from the included study we identified, but we did not find evidence of adverse effects of genotyping.
Data collection and analysis
We included a 'Summary of findings' table in our review which summarised our primary and secondary outcomes for the comparison of genetic testing versus no genetic testing.
Selection of studies
Two review authors (AA and AJ) independently assessed all the titles and abstracts of publications identified by the searches to assess their eligibility. The full texts of all papers found to be eligible at this initial stage were then assessed for inclusion. Publications found to be irrelevant after reading their full text were excluded, and 'Characteristics of excluded studies' tables were prepared to identify the reasons for exclusion. Consensus on the final list of trials to include was reached by discussion.
Data extraction and management
Two review authors (AA and AJ) independently extracted data from the included studies and resolved disagreements by discussion with a third author (TC). The following information on study characteristics and methods were collected into a standardised data extraction form:
-
study design;
-
inclusion and exclusion criteria;
-
setting;
-
country;
-
language of publication;
-
ethnicity of participants;
-
control population (comprising individuals exposed to the culprit drug with no adverse effects or individuals from a healthy population who were not exposed to medications used in the trial);
-
description of genotyping techniques used;
-
genotyping quality control, which included deviation from Hardy‐Weinberg equilibrium (Hardy‐Weinberg equilibrium is a crucial concept in population genetics; it predicts how gene frequencies will be inherited from generation to generation and is used as a measure of quality of genetic tests) and genotype call rate;
-
age;
-
gender;
-
concomitant medications;
-
time from exposure to the culprit drug to skin reaction;
-
type of skin reaction;
-
location of skin lesion;
-
sequelae of adverse reactions;
-
other manifestations indicating systemic involvement; and
-
clinical laboratory tests.
No language translations of papers were required.
Assessment of risk of bias in included studies
Two review authors (AA and AJ) independently assessed the risk of bias in each trial. We used Cochrane's tool for assessing risk of bias (Table 8.5.a in the Cochrane Handbook for Systematic Reviews of Interventions), which is based on seven domains (Higgins 2011):
-
random sequence generation (selection bias);
-
allocation concealment (selection bias);
-
blinding of participants and personnel (performance bias);
-
blinding of outcome assessment (detection bias);
-
incomplete outcome data (attrition bias);
-
selective outcome reporting (reporting bias); and
-
other risk of bias.
The tool allows for the risk of bias for each domain to be assessed as 'low', 'high', or 'unclear' (indicating lack of information or uncertainty over the potential for bias). An additional review author (MP) was consulted in the case of disagreements.
Measures of treatment effect
We planned to use statistical methods in accordance with theCochrane Handbook for Systematic Reviews of Interventions (Higgins 2011). We planned to calculate mean difference (MD) with 95% confidence interval (CI) for continuous data or indeed standardised mean difference (SMD) with 95% CI where all studies reported an outcome using different but similar scales. A risk ratio (RR) with 95% CI was calculated for dichotomous data.
Unit of analysis issues
There were no unit of analysis issues. If such issues arise in an update of this review, we plan to take the appropriate approach to analysis to avoid a unit of analysis error due to repeated observations on participants, multiple treatments, or re‐occurring events. In future updates, if we include cluster‐randomised trials, we will take into account issues such as recruitment bias, baseline comparability of clusters, and number of clusters, and make sure that appropriate statistical methods are used that take into account weighting, etc. (Higgins 2011).
Dealing with missing data
We considered the possible different types of missing data. We planned to deal with missing studies (and the associated risk of bias) by assessing for publication bias, and to deal with missing outcomes (and the associated risk of bias) by assessing for selective reporting (see Assessment of reporting biases). However this was not applicable, due to there only being one included study in this review.
The included study did not report on either of our primary outcomes, but instead reported on an outcome of hypersensitivity reaction which included (but was not limited to) our secondary outcomes HSS and SJS/TEN. So in order to deal with missing data we contacted the study author for additional data on the outcomes of our review. We did not receive a response and therefore we are unaware if the outcomes were measured, and were unable to include these data in our review.
Assessment of heterogeneity
Had a meta‐analysis been possible, we planned to assess the extent of heterogeneity using the I² statistic. We would have used the following thresholds for the interpretation of the I² statistic:
-
0% to 40% = might not be important;
-
30% to 60% = moderate heterogeneity;
-
50% to 90% = may represent substantial heterogeneity; and
-
75% to 100% = considerable heterogeneity.
Due to there being only one included study, we did not need to assess heterogeneity. If eligible studies become available in the future during an update of this review, we will perform a meta‐analysis, report the I² statistic, and interpret the two data together.
Assessment of reporting biases
We planned to assess publication bias by using a funnel plot, Begg test and Egger test (Begg 1989; Egger 1998). Tests for funnel plot asymmetry would only be undertaken when at least 10 studies could be included in the meta‐analysis. Due to the fact that there was only one included study, the power of the tests is too low to distinguish chance from real asymmetry; therefore, this was not assessed.
Data synthesis
Had there been sufficient studies and no significant clinical heterogeneity, we planned to synthesise the results in meta‐analyses, stratified according to type of intervention (e.g. type of genetic test). A random‐effects model would have been assumed.
Where events are rare, a random‐effects approach may be inappropriate. Where events were rare, we planned to take extra care to adopt appropriate methods of meta‐analysis, as recommended in the Cochrane Handbook for Systematic Reviews of Interventions (section 16.9) (Higgins 2011), since many meta‐analysis methods are suboptimal where events are rare through results being biased, confidence intervals being too wide, or power being too low. Choice of method would have been guided by control group risk, likely treatment effect size, and consideration of balance in numbers of treated and control participants in the constituent studies. Had the control groups differed — e.g. if they were drawn from a healthy population or from a population of people treated with the culprit drug but without any adverse effects — we planned to conduct separate analyses. We planned to use Review Manager 5 software to undertake the meta‐analyses (Review Manager 2014).
As we only included one study, it was not possible to undertake a meta‐analysis.
Subgroup analysis and investigation of heterogeneity
We planned to assess heterogeneity using the I² statistic and in the event of substantial heterogeneity, we planned to explore the causes by way of subgroup analyses. We planned to consider the following:
-
participant factors (age, ethnicity, classification of adverse drug reactions, and comparability of participant groups); and
-
trial design issues (genotyping methodology and quality control, blinding, drugs included, and drug dosage and duration of use).
As we only included one study, it was not possible to assess for heterogeneity and therefore a subgroup analysis was not relevant.
Sensitivity analysis
We planned to evaluate the robustness of the results of the meta‐analyses by removing trials of low methodological quality as identified by their risk of bias. As we only included one study, it was not possible to undertake a meta‐analysis, or indeed sensitivity analyses.
Results
Description of studies
Results of the search
The searches of the five databases (see Electronic searches) retrieved 1790 records. Our checks of the trials registers and bibliographies of included studies and relevant reviews did not identify any further relevant studies. Once duplicates were removed we had a total of 1786 records. We excluded 1774 records based on titles and abstracts. We obtained the full text of the remaining 12 records. We excluded nine studies (see Characteristics of excluded studies). One study is awaiting classification (see Characteristics of studies awaiting classification). We did not identify any ongoing studies. We included one study (Mallal 2008), which was reported in two papers.
For a further description of our screening process, see the study flow diagram in Figure 2.
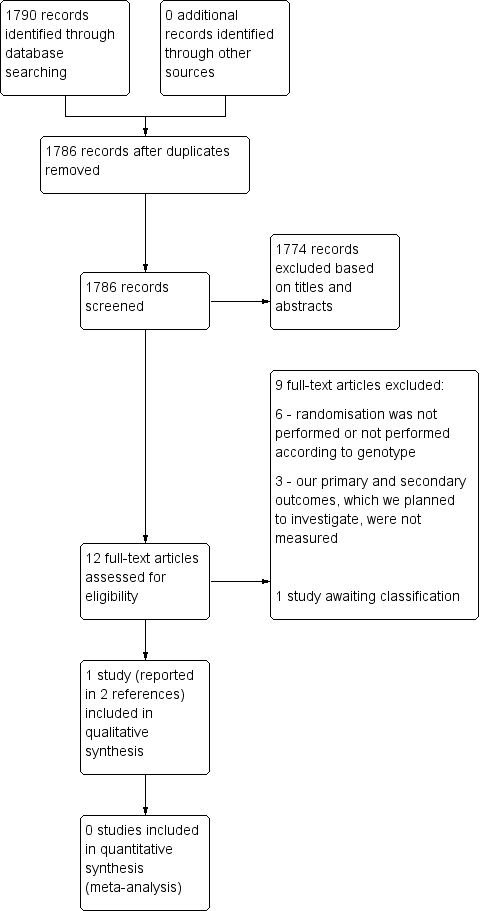
Study flow diagram.
Included studies
Only one study fulfilled our inclusion criteria (Mallal 2008). The summary information can be found in Characteristics of included studies.
This study was a randomised, prospective, double‐blind, multicentre controlled trial, which included 1956 participants from 265 centres (medical centres, hospitals, outpatient clinics) in 19 countries. The participants were infected with HIV‐type 1 (HIV‐1) and had not previously been exposed to the antiretroviral drug abacavir. The study population consisted of men and women (74% men) between the ages of 18 and 77 years old (mean age: 42 years) who were predominantly white. The initial target sample size was 1578 participants. The study was supported by GlaxoSmithKline.
Participants were randomly assigned to undergo prospective pharmacogenetic screening for the human leukocyte antigen class I gene HLA‐B*57:01 (prospective screening group, i.e. the treatment group) or to undergo a standard care approach without genetic testing (control group). In the prospective screening group, HLA‐B*57:01‐positive participants were excluded from abacavir treatment to prevent skin rash. These individuals were given a combination of active antiretroviral therapy that did not include abacavir. (Those that tested negative for HLA‐B*57:01 were given a combination of active antiretroviral therapy including abacavir.)
All those in the control group were given a combination of active antiretroviral therapy including abacavir without prospective screening for HLA‐B*57:01. These participants had retrospective HLA‐B*57:01 pharmacogenetic testing.
The study encompassed six‐week observation periods conducted over a six‐month trial period from April to September 2006.
The primary outcomes reported by the study were to investigate whether there was a reduction in clinically diagnosed hypersensitivity reactions to abacavir in the genotyped group compared to the control group, and to explore the rate of immunologically confirmed hypersensitivity reaction (defined as "clinically diagnosed reaction that was confirmed by a positive result on epicutaneous patch testing 6 to 10 weeks after clinical diagnosis" (Mallal 2008)). Assessment of hypersensitivity was performed at the time of study entry at baseline (day one) and at weeks one, two and six. None of the review's primary outcomes were reported in this trial. In terms of the review's secondary outcomes, HSS and SJS/TEN were reported, but only as a combined outcome under 'hypersensitivity reactions'.
Excluded studies
We excluded nine studies following full‐text review. Reasons for exclusion were as follows:
-
randomisation was not performed, or not performed according to genotype (Bonnefoi 2011; Cheng 2009; Pusztai 2014; Sequist 2013; Seymour 2013; Young 2008); and
-
our primary and secondary outcomes, which we planned to investigate, were not measured (Azuma 2013; Damronglerd 2015; Newman 2011).
Full reasons for exclusion are listed in Characteristics of excluded studies.
Risk of bias in included studies
The overall risk of bias was low, with the exception of a notably high risk of attrition and detection bias (Figure 3; Figure 4).
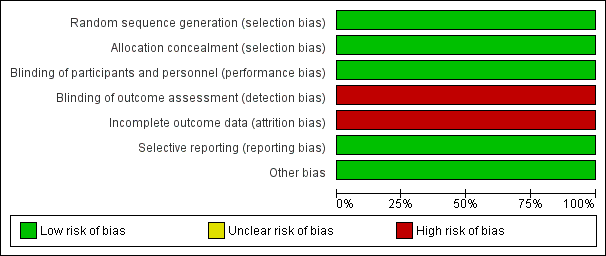
Risk of bias graph: review authors' judgements about each risk of bias item presented as percentages across all included studies.

Risk of bias summary: review authors' judgements about each risk of bias item for each included study.
Allocation
Random sequence generation
All 1956 participants in the study were adequately randomised based on a computer‐generated, centralised schedule; thus, allocation to the treatment and control groups (prospective and retrospective testing) indicates a low risk of selection bias.
Allocation concealment
The method of allocation concealment represents low risk of bias because it was performed by the central study‐management group without foreknowledge of intervention assignments. The sequence was implemented in the block sizes of four. Stratification was done according to ethnicity to ensure a good balance of ethnic subgroups between the two trial arms, since risk of outcomes can vary across ethnic groups.
Blinding
Blinding was undertaken sufficiently, with the investigators, participants and study management team remaining unaware of the participant assignments to the treatment or control group. Therefore we deemed the study to have a low risk of performance bias. In terms of detection bias, the assessors were unaware of the genetic test results and investigators were trained using illustrated guides and an informational video. However, hypersensitivity reactions were diagnosed by the principal investigator at the recruitment site without the use of predefined clinical criteria and this poses a risk of detection bias.
Incomplete outcome data
Participants treated with abacavir, but for whom there were incomplete follow‐up data for various detailed reasons, were excluded from the primary analyses; therefore these analyses are at risk of attrition bias. However, potential bias introduced by the exclusion of the participants who could not be evaluated was addressed by several sensitivity analyses of data from the full intention‐to‐treat population that had received abacavir, with assumptions about the missing data ranging from 0% to 100% of the exclusions being associated with a hypersensitivity reaction. The results of the sensitivity analyses were in line with the results of the primary analyses.
Selective reporting
We judged the study to be at low risk of reporting bias as both prespecified outcomes from the protocol (ClinicalTrials.gov identifier: NCT00340080) were reported. It was not possible to assess for between‐study selective reporting as there was only one included study.
Other potential sources of bias
No other potential sources of bias appear within the study.
Effects of interventions
See: Summary of findings for the main comparison
The study did not report on either of our primary outcomes (the incidence of severe drug‐induced skin rash; or long‐term sequelae). The study did report 'hypersensitivity reactions' which included, but was not limited to, our secondary outcomes HSS (hypersensitivity syndrome) and SJS/TEN (Stevens‐Johnson syndrome, toxic epidermal necrolysis).
The study demonstrated that prospective HLA‐B*57:01 screening can reduce the incidence of hypersensitivity reaction to abacavir. The incidence of immunologically confirmed hypersensitivity reaction to abacavir was lower in the screening arm (risk ratio 0.02; 95% CI 0.00 to 0.37; P < 0.001; 1644 participants; Analysis 1.1), as was the incidence of clinically diagnosed hypersensitivity reaction to abacavir (risk ratio 0.43; 95% CI 0.28 to 0.67; P < 0.001; 1650 participants; Analysis 2.1). These results are presented in summary of findings Table for the main comparison, Figure 5 and Figure 6.

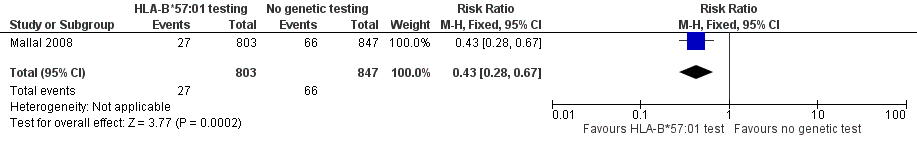
Forest plot of comparison: 1 Genetic testing vs no testing, outcome: 1.1 Hypersensitivity (HSS).

Forest plot of comparison: 1 Genetic testing with skin patch testing versus no genetic testing with skin patch testing, outcome: 1.1 Hypersensitivity (HSS) immunologically confirmed.
However, we are aware of some potential long‐term effects of abacavir. In order to determine any further effects of the intervention, we made an attempt to contact the study authors for information on our remaining primary and secondary outcomes (as mentioned above in Dealing with missing data). We did not receive a response, and therefore are unable to estimate any further effects of the intervention.
Discussion
Summary of main results
Our one included study (with a total of 1956 participants) did not report on either of our primary outcomes of interest, which were:
-
the incidence of severe drug‐induced skin rash (defined as skin rash with systemic symptoms including fever and multiple organ involvement); and
-
long‐term sequelae (any of the following: ophthalmologic, cutaneous, or liver damage, etc.) up to 12 months after the severe drug‐induced skin rash.
The study addressed a combination of our second and fourth secondary outcomes, which were: SJS/TEN (Stevens‐Johnson syndrome, toxic epidermal necrolysis) and HSS (hypersensitivity syndrome). Based on moderate‐quality evidence, prospective HLA‐B*57:01 screening probably reduces the incidence of hypersensitivity reaction to abacavir compared to when prospective pharmacogenetic screening is not performed. However, hypersensitivity reactions were diagnosed by the principal investigator at the recruitment site without the use of predefined clinical criteria and therefore there is a high risk of detection bias.
Carriers of HLA‐B*57:01 were excluded from receiving abacavir; the rate of clinical diagnosis of hypersensitivity reaction was 7.8% in the control group (who received no screening, but underwent HLA typing retrospectively after abacavir exposure) compared to 3.4% (95% CI 2.2% to 5.2%) in the prospective‐screening group (risk ratio (RR) 0.43, 95% CI 0.28 to 0.67) (summary of findings Table for the main comparison; Figure 5).
In addition, undertaking skin patch testing to confirm the diagnosis of hypersensitivity and excluding carriers of HLA‐B*57:01 from receiving abacavir, reduced the rate of immunologically confirmed diagnosis of hypersensitivity from 2.7% in the control group (no screening) to zero in the prospective‐screening group (RR 0.02, 95% CI 0.00 to 0.37) (summary of findings Table for the main comparison; Figure 6).
Based on this trial, pretreatment genetic testing for the HLA‐B*57:01 allele has been recommended by the Clinical Pharmacogenetics Implementation Consortium (Martin 2014) and several regulating agencies, including the Food and Drug Administration (FDA), European Medicines Agency (EMA) and Pharmaceuticals and Medical Devices Agency (PMDA).
Overall completeness and applicability of evidence
The included study is not sufficient to address all the objectives of this review (Mallal 2008). Whilst it did provide some data related to two secondary outcomes, and was generally at low risk of bias, more studies are required if our objectives are to be explored fully. The study is the only randomised controlled trial that assessed efficacy and safety of genetic testing in prediction of adverse drug reactions. Included individuals were representative of the study population of interest. However, it will be important in future studies to explore ethnic variability as this study has been conducted in a predominantly white population. The follow‐up period was short and there was no information on long‐term sequelae in drug‐hypersensitivity survivors. The study did not report on either of our primary outcomes of interest. In addition, information on hospitalisation, AGEP (acute generalised exanthematous pustulosis), or death were not reported either. We found no eligible evidence with regard to genetic testing for severe drug‐induced skin rash in relation to different drugs and classes of drugs.
Quality of the evidence
The study included in this review has adequate randomisation methods, as well as methods of allocation concealment. However, the included trial did not report on the characteristics of the 55 participants (5.6%) who were positive for HLA‐B*57:01 and therefore did not receive abacavir treatment and were excluded from data analysis. Information on outcomes within this patient group would have been informative. There is also potential for detection bias as the principal investigator diagnosed hypersensitivity reactions without the use of predefined clinical criteria. Therefore, we downgraded the quality of evidence by one level due to study limitations (risk of bias). We did not find any reason to downgrade the evidence for the other GRADE domains (inconsistency, imprecision, indirectness, or publication bias). Therefore, quality of evidence was rated as moderate using the GRADE criteria (Schünemann 2013).
Potential biases in the review process
We did not identify any sources of potential bias in the review process. We carefully assessed diverse terminology used to describe drug‐induced hypersensitivity and included alternative search terms in our literature searches. We clearly defined the outcomes and patient subgroups. As we included only RCTs in our protocol, no other study designs were considered. We did not impose any date restrictions on the search. We contacted the study authors to provide additional data, but that did not generate any additional information.
Agreements and disagreements with other studies or reviews
To our knowledge there have been no previous systematic reviews addressing this research question, and the only previous study addressing the research question is included as the only study in this review (Mallal 2008). Due to there only being one identified RCT, there are remaining uncertainties about the effects of prospective pharmacogenetic screening to reduce drug‐associated skin reactions in a patient population. Several studies have recently reported clinical utility of pretreatment genetic testing that can predict and prevent serious cutaneous adverse drug reactions (Chen 2011; Fang 2019; Liu 2019; Mushiroda 2018; Park 2019; Stainsby 2019). However, these studies are not RCTs and therefore were not included in our review; they use historical frequency data on adverse drug reactions over a period of several years to control for HLA allele screening that helps to predict and prevent adverse drug reactions in carriers of risk alleles. The findings in these studies agree with the conclusions of our review.

Flowchart of interventions (genetic testing) and outcomes (skin rash) in a patient population prescribed drug X
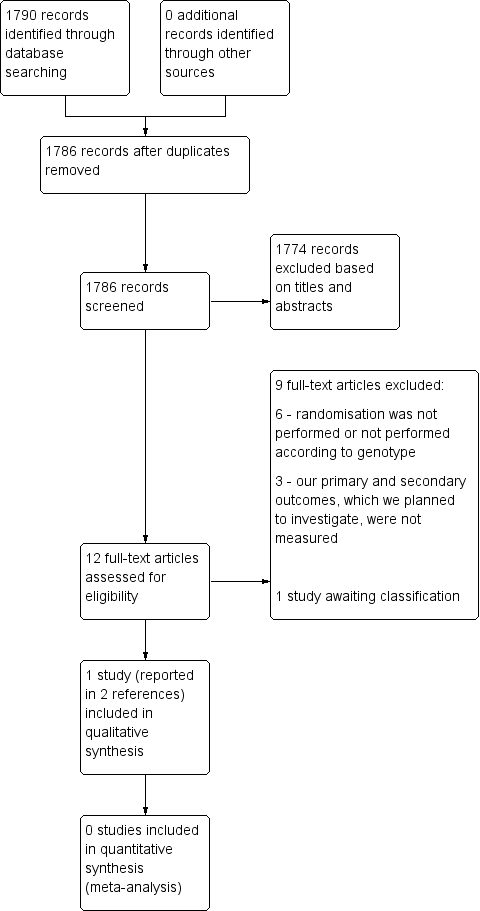
Study flow diagram.

Risk of bias graph: review authors' judgements about each risk of bias item presented as percentages across all included studies.

Risk of bias summary: review authors' judgements about each risk of bias item for each included study.
Forest plot of comparison: 1 Genetic testing vs no testing, outcome: 1.1 Hypersensitivity (HSS).

Forest plot of comparison: 1 Genetic testing with skin patch testing versus no genetic testing with skin patch testing, outcome: 1.1 Hypersensitivity (HSS) immunologically confirmed.

Drug‐induced skin rash: top panel = maculopapular exanthema, bottom panel = Steven Johnson Syndrome (blistering skin rash with skin detachment)
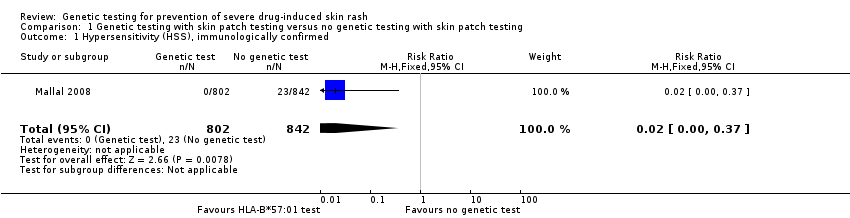
Comparison 1 Genetic testing with skin patch testing versus no genetic testing with skin patch testing, Outcome 1 Hypersensitivity (HSS), immunologically confirmed.
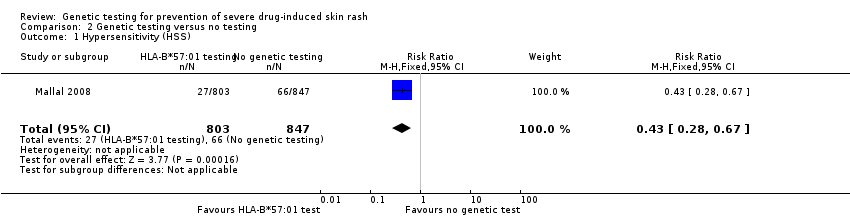
Comparison 2 Genetic testing versus no testing, Outcome 1 Hypersensitivity (HSS).
| Prospective genetic HLA‐B*57:01 screening compared with standard care for drug‐induced skin rash | ||||||
| Patient or population: patients with HIV‐1 infection and a pre‐established clinical need for treatment with an antiretroviral drug regimen containing abacavir but with an unknown HLA‐B*57:01 status. Settings: secondary care clinics Intervention: prospective genetic screening for the HLA‐B*57:01 allele Comparison: no prospective genotyping, standard‐of‐care treatment | ||||||
| Outcomes | Illustrative comparative risks* (95% CI) | Relative effect | No of Participants | Quality of the evidence | Comments | |
| Assumed risk | Corresponding risk | |||||
| Standard Carea | Prospective genetic HLA‐B*57:01 screening | |||||
| Severe skin drug‐induced rash | No data | ‐ | ‐ | ‐ | Not assessed | |
| Long‐term sequelae | No data | ‐ | ‐ | ‐ | Not assessed | |
| Hospitalisation for drug‐induced skin reaction | No data | ‐ | ‐ | ‐ | Not assessed | |
| SJS/TEN (Stevens‐Johnson syndrome/toxic epidermal necrolysis) | See hypersensitivity | ‐ | ‐ | ‐ | Not assessable | |
| AGEP (acute generalised exanthematous pustulosis) | No data | ‐ | ‐ | ‐ | Not assessed | |
| HSS (hypersensitivity) reaction including SJS/TEN (clinically diagnosed) (6 weeks clinical assessment) | 78 per 1000 | 34 per 1000 | RR 0.43 (0.28 to 0.67) | 1650 participants | ⊕⊕⊕⊝ | ‐ |
| HSS reaction including SJS/TEN (immunologically confirmed) (6 weeks clinical assessment) | 27 per 1000 | 0 per 1000 | RR 0.02 (0.00 to 0.37) | 1644 participants (1 study) | ⊕⊕⊕⊝ moderateb | ‐ |
| Death | No data | ‐ | ‐ | ‐ | Not assessed | |
| *The basis for the assumed risk (e.g. the median control group risk across studies) is provided in footnotes. The corresponding risk (and its 95% confidence interval) is based on the assumed risk in the comparison group and the relative effect of the intervention (and its 95% CI). | ||||||
| GRADE Working Group grades of evidence | ||||||
| a The assumed risk is estimated from the event rate (confirmed clinically diagnosed occurrences of HSS including SJS/TEN or immunologically confirmed) in the control arm of the included study. b We downgraded the evidence to moderate quality, due to study limitations (high risk of detection and attrition bias). | ||||||
| Term | Explanation |
| Allele | One of two or more alternative forms of a gene at corresponding sites (loci) on homologous chromosomes |
| Antiretroviral | A class of drugs that inhibit the activity of retroviruses that cause HIV infection |
| Hardy‐Weinberg equilibrium | This states that allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences |
| HLA | Human leukocyte antigen: a group of protein molecules located on bone marrow and other cells that can provoke an immune response |
| Hypersensitivity | A state of altered reactivity in which the body reacts with an exaggerated immune response to a foreign substance, such as a drug |
| Immunologically confirmed | Patch testing is done to see whether a particular drug is causing allergic skin reaction. Patch test can detect delayed allergic or immunological reaction and confirm the diagnosis of hypersensitivity. |
| Phenotypes | The set of observable characteristics of an individual resulting from the interaction of its genotype with the environment |
| Polymorphic | A variation in the DNA that is too common to be due merely to new mutation. A polymorphism must have a frequency of at least 1% in a population |
| Maculopapular rash | A rash with both macules (flat and coloured like a freckle) and papules (a small raised spot) |
| Sequelae | A condition that is a consequence of a previous disease or injury |
| T‐cells | Another term for T‐lymphocyte, a type of cell that participates in immune response |
| Drugs associated with skin injury | Class of drug | HLA allele | Population | Reference |
| Stevens‐Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) | ||||
| Allopurinol | Antiuric acid | B*5801 | Han Chinese | |
| ‐ | ‐ | ‐ | Thai | |
| ‐ | ‐ | ‐ | Japanese | |
| ‐ | ‐ | ‐ | Malay | |
| Carbamazepine | Antiepileptic | B*1502 | Han Chinese | |
| ‐ | ‐ | ‐ | Thai | Kulkantrakorn 2012; Locharernkul 2008; Tassaneeyakul 2010; Tangamornsuksan 2013 |
| ‐ | ‐ | ‐ | Malay | |
| ‐ | ‐ | ‐ | Indian | |
| ‐ | ‐ | A*3101 | White | |
| ‐ | ‐ | A*3101 | Japanese | |
| Phenytoin | Antiepileptic | B*1502 | Han Chinese | |
| ‐ | ‐ | ‐ | Thai | |
| Oxicam | Non‐steroidal anti‐inflammatory drug (NSAID) | A2, B12 | White | |
| Sulphamethoxazole | Antibiotic | A29, B12, DR7 | White | |
| Hypersensitivity syndrome (drug‐induced hypersensitivity syndrome (DIHS) or drug reaction with eosinophilia and systemic symptoms (DRESS)) | ||||
| Abacavir | Antiretroviral | B*57:01 | White | Hetherington 2002; Hughes 2004a; Mallal 2002; Mallal 2008; Martin 2004 |
| ‐ | ‐ | ‐ | African American | |
| Aminopenicillins | Antibiotic | A2, Drw52 | White | |
| Nevirapine | Antiretroviral | DRB1*01 | White ‐ Australian | |
| ‐ | ‐ | DRB1*01 | White ‐ French | |
| ‐ | ‐ | Cw8, B14 | White ‐ Italian | |
| ‐ | ‐ | Cw8 | Japanese | |
| ‐ | ‐ | B*3505 | Thai | |
| ‐ | ‐ | Cw4 | Thai | |
| ‐ | ‐ | C*0404 | Black African | |
| ‐ | ‐ | Cw*04 | Chinese | |
| Aspirin | NSAIDs | DRB1*1302, DQB1*0609 | ‐ | |
| ‐ | NSAIDs | DR11 | ‐ | |
| Iodine contrast media | ‐ | DR | White ‐ Spanish | |
| Paraphenylenediamine | Hair dye | DP | White ‐ German | |
| Gold sodium thiomalate | Treatment of rheumatoid arthritis | DR5 | White ‐ Spanish | |
| Lamotrigine | Antiepileptic | B*5801, A*6801 | White | |
| Trichloroethylene | Industrial solvent, dry cleaning | B*1301 | Japanese | |
| Fixed drug eruptions | ||||
| Co‐trimoxazole | Antibiotic | A30, B13, Cw6 | White ‐ Turkish | |
| Feprazone | Analgesic | B22 | ‐ | |
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
| 1 Hypersensitivity (HSS), immunologically confirmed Show forest plot | 1 | 1644 | Risk Ratio (M‐H, Fixed, 95% CI) | 0.02 [0.00, 0.37] |
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
| 1 Hypersensitivity (HSS) Show forest plot | 1 | 1650 | Risk Ratio (M‐H, Fixed, 95% CI) | 0.43 [0.28, 0.67] |

