Molecular assays for the diagnosis of sepsis in neonates
Referencias
References to studies included in this review
References to studies excluded from this review
Additional references
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Ir a:
| Study characteristics | |||
| Patient sampling | Participant sampling not clearly described. | ||
| Patient characteristics and setting | Newborns > 3 days old with suspected sepsis. No information on participant demographics or study period. | ||
| Index tests | PCR using universal candida DNA sequence. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples drawn at the same time. | ||
| Comparative | |||
| Notes | Data from conference abstract only. No information on participant demographics or study period. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Participants were recruited consecutively. | ||
| Patient characteristics and setting | Preterm infants < 37 weeks and > 72 hours old with signs and symptoms of sepsis requiring antibiotic treatment. Interquartile range of age reported suggests some infants may have been > 28 days of age. Study period: March 2006 to June 2008 (28 months). | ||
| Index tests | Real‐time PCR using universal primers and Gram‐specific probes. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood, peritoneal fluid and CSF cultures. | ||
| Flow and timing | Index test and the reference standard performed at the same time. | ||
| Comparative | |||
| Notes | 15 samples were excluded due to insufficient amount of sample (n = 9) and mistakenly left in the refrigerator for > 72 hours (n = 6). Excluded samples not included in the analysis. Cycle threshold cut‐off values for positive PCR were defined. Interquartile range of age reported suggests some infants may have been > 28 days of age. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study did not classify whether participants were enrolled randomly or consecutively. Negative controls (n = 30) were not included in the analysis. | ||
| Patient characteristics and setting | Infants with suspected sepsis, admitted to the neonatal department and the intensive care unit of the Zhejiang University Children's University in China. It was unclear how many infants were < 28 days old as no participant demographics are available. Study period: September 2007 to June 2008. | ||
| Index tests | Broad‐range 16S rRNA‐based real‐time fluorescent PCR. | ||
| Target condition and reference standard(s) | Suspected sepsis and the reference standard were cultures of blood and CSF. | ||
| Flow and timing | Both index test and reference standard samples were drawn simultaneously. | ||
| Comparative | |||
| Notes | No participant demographics available and unclear if some infants were > 28 days of age. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | All neonates with suspected sepsis admitted during the period of May 2012 to August 2012 were enrolled. | ||
| Patient characteristics and setting | Neonates with suspected sepsis admitted to the NICU of Ain Shams University Hospitals. Study period: May 2012 to August 2012. Age range reported was 0 to 50 days. | ||
| Index tests | Broad‐range 16S rDNA PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood sample for culture and PCR were collected concurrently using standard sterile procedures. | ||
| Comparative | |||
| Notes | Participants were referred to as neonates although the age range reported was 0 to 50 days. Participants included both preterm and full‐term infants. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Not clearly reported. | ||
| Patient characteristics and setting | Neonates with suspected sepsis admitted to Level III NICU. Study period not mentioned. | ||
| Index tests | Broad‐range conventional PCR after 5‐hour preamplification culture. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for culture and PCR were drawn simultaneously. Reason for exclusion of participants were reported. | ||
| Comparative | |||
| Notes | Of the 64 participants that were excluded, 34 had malformations, 15 had < 12‐hour life expectancy and the remaining 15 had contaminated blood cultures. Study period not mentioned. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met the inclusion criteria during the study period. Controls (n = 50) were not included in the analysis. | ||
| Patient characteristics and setting | Newborn participants with signs and history suggestive of sepsis admitted in the NICU at Kobe Hospital University from June 2005 to September 2006. | ||
| Index tests | Multiplex PCR targeting 8 common pathogens. | ||
| Target condition and reference standard(s) | Neonatal sepsis and bacterial culture of blood, skin, bronchoalveolar lavage, mucus, CSF, urine and ascitic fluid. | ||
| Flow and timing | Only 77 samples with paired specimen culture and PCR were included in the 2 × 2 table. Samples for culture and PCR were drawn simultaneously. | ||
| Comparative | |||
| Notes | Of the 6 specimens that were positive for PCR but negative for culture, 1 culture was positive for normal flora and was considered negative. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met the inclusion criteria during the study period. | ||
| Patient characteristics and setting | Newborns < 7 days old with suspected sepsis or meningitis diagnosed at a participating hospital from November 2005 to January 2007. | ||
| Index tests | RT‐PCR targeting the 16S rRNA. | ||
| Target condition and reference standard(s) | Suspected early‐onset neonatal sepsis and blood and CSF cultures. | ||
| Flow and timing | Sample for PCR and culture were drawn concurrently. Samples for PCR were stored until DNA extraction. | ||
| Comparative | |||
| Notes | Analyzed only EOS in neonates and included 83 neonates. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Neonates admitted to the NICU of Jutendo University Hospital or Jutendo Shizuoka Hospital from February to August 2009. Mean (SD) gestational age was 34.8 ± 5.8 weeks. There were 36 participants with 39 episodes of sepsis. | ||
| Index tests | RT‐PCR targeting 16S rRNA. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Whole blood collected concurrently for PCR and culture. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Neonates up to 28 days old admitted to the NICU from August 2005 to July 2006. | ||
| Index tests | Broad‐range PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Index test and reference standard sampling performed simultaneously. | ||
| Comparative | |||
| Notes | Only blood culture‐positive samples were included in the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| High | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Participants who met the inclusion criteria were enrolled prospectively. | ||
| Patient characteristics and setting | Neonates with suspected clinical sepsis admitted to the Central South Hospital of Petroleos Mexicanos, the Gynecological‐Obstetrics Hospital number 4 of the Mexican Institute of Social Security, the Dalinde Hospital and the Monterrey Nuevo Leon University Hospital and National Institute of Perinatology. Study period not mentioned. | ||
| Index tests | LightCycler SeptiFast Test. | ||
| Target condition and reference standard(s) | Suspected neonatal sepsis and blood culture. | ||
| Flow and timing | Samples for blood culture and LightCycler SeptiFast were drawn concurrently. | ||
| Comparative | |||
| Notes | Study period not mentioned in the report. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | All infants admitted to the NICU for sepsis evaluation. | ||
| Patient characteristics and setting | All infants admitted to the NICU for sepsis evaluation. No participant demographics available. | ||
| Index tests | Broad‐range conventional PCR and DNA dot‐blot hybridization. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Index test and reference standard were performed simultaneously. | ||
| Comparative | |||
| Notes | This was a feasibility study and blood sample for PCR was from discarded or unused sample sent to evaluate CBCs. It was not clear whether blood drawn for CBC was also done with the same aseptic technique as blood culture. Study period not mentioned. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met inclusion criteria. | ||
| Patient characteristics and setting | Infant admitted to the NICU for sepsis evaluation that included at least blood culture and CBC. No demographic information or study period details available. | ||
| Index tests | Real‐time 16S rRNA PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood sample used for PCR was from discarded or unused samples sent for evaluation of CBC. Unclear whether blood drawn for CBC was done in an aseptic manner. | ||
| Comparative | |||
| Notes | Study was done to design a sample preparation protocol that would eliminate tryptic soy broth pre‐enrichment step and to convert conventional PCR assay to a real‐time PCR platform. The methodology here is real‐time PCR from whole blood without enrichment. So a different methodology fromJordan 2000paper and overlap is very unlikely. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | All NICU admissions during the period of study were screened for eligibility. | ||
| Patient characteristics and setting | Infants > 34 weeks admitted to the NICU for suspected EOS from 1 September 2000 to 1 April 2004. | ||
| Index tests | Broad‐range conventional PCR followed by pyrosequencing. | ||
| Target condition and reference standard(s) | EOS in near‐term infants and blood culture. | ||
| Flow and timing | Samples for the index test and reference standard were collected simultaneously but PCR was evaluated from sample sent for CBC. Concerns about aseptic technique remain. | ||
| Comparative | |||
| Notes | Blood samples for PCR were from unused portion of the sample sent to evaluate CBC and were collected by venous, arteria or heel stick. The PCR was conventional PCR with enrichment with Trypticose soy before PCR just like the paperJordan 2000. The study period here was stated to be from September 2000.Jordan 2000paper was submitted for publication in 1999 as per the title page of the article and hence overlap ofJordan 2000andJordan 2006unlikely. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | Neonates who met inclusion criteria were enrolled on admission. | ||
| Patient characteristics and setting | VLBW infants > 72 hours old. Participant demographics or study period not available. | ||
| Index tests | Multiplex real‐time PCR using Roche LightCycler SeptiFast MGRADE system. | ||
| Target condition and reference standard(s) | Neonates with suspected LOS and blood culture. | ||
| Flow and timing | Blood sample for PCR was collected during routine sepsis work‐up and before antibiotics. | ||
| Comparative | |||
| Notes | Participant demographics or study period not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Newborn at risk for EOS from January to September 1996. Predefined major and minor criteria were used to classify participants "at risk" for sepsis. | ||
| Index tests | Broad‐range conventional PCR | ||
| Target condition and reference standard(s) | Neonatal EOS and blood culture. | ||
| Flow and timing | Blood samples for analyses were drawn concurrently. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Neonates were enrolled if they met inclusion criteria during the study period. | ||
| Patient characteristics and setting | Neonates with suspected sepsis during the period of December 2004 to June 2005. Participant demographics not available. | ||
| Index tests | Real‐time PCR using universal primers. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | Recalculated sensitivity, specificity, PPV, and NPV as samples positive for PCR were also positive for human DNA and not bacterial DNA. Participant demographics not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | All neonates with suspected sepsis and had blood samples drawn for concomitant culture, CBC and CRP assay were included in the study. | ||
| Patient characteristics and setting | Neonates with suspected sepsis admitted to the NICU of the Women and Children's Hospital, the Children's Hospital and Tongji Hospital in Hubei Province from 1 September 2011 to 31 December 2011. Participants were from 4 hour to 28 days old. | ||
| Index tests | 16S rRNA gene PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Additional 0.5 mL to 1 mL EDTA blood sample was collected for PCR at the time of sepsis workup. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Prospective enrollment of infants that met inclusion criteria during a 12‐month period. | ||
| Patient characteristics and setting | Neonates aged > 3 days, admitted to the NICU with suspected LOS. Gestational age range 24 to 42 weeks and range of age at enrollment was 4 to 96 days. Study period not mentioned although reported over 12 months. | ||
| Index tests | Staphylococcal 16S rRNA PCR (both Staphylococcus aureus and coagulase‐negative Staphylococcus). | ||
| Target condition and reference standard(s) | Neonatal LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | There were 32 culture‐positive samples for bacteria and fungi but only 13 were positive for staphylococci and this was incorporated into the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Prospective enrollment of neonates that met the criteria for suspected LOS. | ||
| Patient characteristics and setting | Neonates aged > 3 days with suspected LOS. The age range of infants included were 4 to 105 days. Study period not available. | ||
| Index tests | Staphylococcal 16S rRNA PCR (both Staphylococcus aureus and coagulase‐negative Staphylococci). | ||
| Target condition and reference standard(s) | Neonates with suspected LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | The article mentioned 148 events of LOS but on further scrutiny there were on 146 events which were incorporated into the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Newborn infant that met inclusion criteria for EOS and LOS admitted to the NICU during the period of 1999 to 2005. | ||
| Patient characteristics and setting | Newborn infants < 28 days old with suspected EOS or LOS admitted to Öbrero University from 1999 to 2005. | ||
| Index tests | Real‐time PCR targeting 16S rRNA. | ||
| Target condition and reference standard(s) | Neonates with suspected EOS or LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn simultaneously. | ||
| Comparative | |||
| Notes | PCR results from 1 sample that was positive for culture and PCR was considered uninterpretable as PCR result showed double sequence. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | All infants that met inclusion criteria were enrolled prospectively. | ||
| Patient characteristics and setting | All infants aged < 3 months who underwent sepsis evaluation and admitted to the NICU at 2 Swedish University Hospitals between October 2007 and November 2009. Of the participants enrolled in the study, 34 infants were > 28 days old. | ||
| Index tests | Broad‐range 16S real‐time PCR. | ||
| Target condition and reference standard(s) | Suspected sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn simultaneously. | ||
| Comparative | |||
| Notes | 16 participants were excluded due to lack of consent, 7 for being older than 3 months and 10 participants whose blood sample for PCR and culture were not drawn concurrently. Excluded participants were not included in the analysis. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | 34 newborns with LOS were enrolled in the study. | ||
| Patient characteristics and setting | Newborns > 3 days old with suspected LOS. Age of participants at enrollment and study period not available. | ||
| Index tests | Commercial real‐time PCR using LightCycler SeptiFast system (multiplex PCR). | ||
| Target condition and reference standard(s) | Neonatal LOS and blood culture. | ||
| Flow and timing | Blood samples for LightCycler SeptiFast and culture were simultaneously. | ||
| Comparative | |||
| Notes | Age of participants at enrollment and study period not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Prospective, non‐randomized enrollment of participants that met inclusion criteria. | ||
| Patient characteristics and setting | Infants with birth weight > 1000 g admitted to the NICU at Akershus University Hospital with suspected sepsis during the first week of life. Age at study enrollment and study period not mentioned. | ||
| Index tests | Broad‐range 16S rRNA PCR followed by sequencing. | ||
| Target condition and reference standard(s) | Suspected neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | PCR samples were stored until analysis. 4 infants were excluded from the study with 3 having incomplete registration and 1 with missing sample. 1 infant in the final analysis ended up with a diagnosis of asphyxia rather than sepsis. Age at study enrollment and study period not mentioned. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Neonates with clinically suspected sepsis. | ||
| Patient characteristics and setting | Neonates with suspected sepsis. The gestational age ranged from 26 to 39 weeks but age at enrollment not mentioned. Study period: October 2010 to December 2012. | ||
| Index tests | 16S rDNA PCR. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for blood culture and PCR were done simultaneously. | ||
| Comparative | |||
| Notes | Age at enrollment not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | All infants that met inclusion criteria during a specified period of time. Controls were excluded from analysis. | ||
| Patient characteristics and setting | All neonates > 3 days old admitted to the neonatal ward or NICU who developed clinical signs of LOS during the period of 1 January 2004 to June 30, 2004. Other participant demographics not available. | ||
| Index tests | Broad‐range 16S rRNA PCR followed by microarray hybridization. | ||
| Target condition and reference standard(s) | Suspected neonatal LOS and blood culture. | ||
| Flow and timing | Unclear whether blood samples for PCR and blood culture were drawn simultaneously. | ||
| Comparative | |||
| Notes | Participant demographics not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Consecutive enrollment of infants (24 were neonates) with signs of systemic inflammatory response syndrome and risk factors for candidemia. | ||
| Patient characteristics and setting | Infants who were admitted to the ICU of 2 pediatric hospital in Sao Paulo State, Brazil over an 18‐month period. Study period (month and year) or participant demographics not available. Author provided results for the 24 neonates. | ||
| Index tests | Multiplex nested PCR with specific primers designed to identify 7 Candida species | ||
| Target condition and reference standard(s) | Candidemia and blood culture. | ||
| Flow and timing | Blood sample for both culture and PCR were done concurrently. | ||
| Comparative | |||
| Notes | Data based on email communication with Dr. Del Negro. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | All infants with suspected sepsis in the NICU and PICU during the study period were considered for inclusion in the study. | ||
| Patient characteristics and setting | Infants admitted in the NICU (n = 46) and PICU (n = 17) with suspected sepsis during the period from November 1999 to November 2000. PCR and blood culture data separately for neonates not available. | ||
| Index tests | Fungal conventional PCR targeting 18S rRNA. | ||
| Target condition and reference standard(s) | Suspected sepsis and blood culture. | ||
| Flow and timing | Excess blood used for culture was used for PCR. | ||
| Comparative | |||
| Notes | PCR and blood culture data separately for neonates not available. It was unclear how many of the infants admitted in the PICU were neonates hence, not all infants may have met the target condition of neonatal sepsis defined in this study. PCR products were analyzed by 2 independent observers blinded to blood culture results and participant information. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study data derived from conference abstract only and hence limited. | ||
| Patient characteristics and setting | Neonates with suspected sepsis. No participant demographics or study period details available. | ||
| Index tests | 16S rRNA‐based PCR followed by hybridization to chips with 18 probes. | ||
| Target condition and reference standard(s) | Infants with suspected sepsis and blood culture. | ||
| Flow and timing | Possible simultaneous sampling for index test and reference standard. | ||
| Comparative | |||
| Notes | Limited information from abstract. No participant demographics or study period details available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Participants who met inclusion criteria were admitted consecutively. | ||
| Patient characteristics and setting | Infants with febrile episodes admitted to the NICU at the Hospital Universitario Virgen de las Nieves. Study period: April 2007 to April 2009. Participants enrolled in the study were both preterm and term infants; however, age of participants at the time of enrollment range from 0 to 151 days old. | ||
| Index tests | LightCycler SeptiFast Assay. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Sample for blood culture and LightCycler SeptiFast assay were collected at the same time. | ||
| Comparative | |||
| Notes | Participants enrolled in the study were both preterm and term infants; however, age of participants at the time of enrollment range from 0 to 151 days old. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Only participants with probable candidiasis were included in the study. | ||
| Patient characteristics and setting | Neonates at high risk for invasive candidiasis from Jan 2009 to Dec 2010. No information on participant demographics available. | ||
| Index tests | Detection of fungal DNA directly from lysis‐centrifugation blood culture. Fungus‐specific universal primer ITS1 and ITS2 were used to amplify 18S rDNA, the adjacent ITS1 and a small portion of the 28S rDNA region. | ||
| Target condition and reference standard(s) | Suspected neonatal candidiasis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture came from the same Isolator 1.5 microbial tubes. | ||
| Comparative | |||
| Notes | No information on participant demographics available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Consecutive enrollment of preterm infants with suspected LOS. | ||
| Patient characteristics and setting | Preterm infants with suspected LOS admitted to the NICU. Participant demographics or study period not mentioned. | ||
| Index tests | Multiplex real‐time PCR assay. | ||
| Target condition and reference standard(s) | LOS in neonates and blood culture. | ||
| Flow and timing | Blood samples for culture and PCR were drawn concurrently. | ||
| Comparative | |||
| Notes | Participant demographics or study period not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Limited information from abstract. | ||
| Patient characteristics and setting | Newborns aged > 3 days with suspected LOS. Participant demographics or study period data not available. | ||
| Index tests | Broad‐range 16S rRNA conventional PCR. | ||
| Target condition and reference standard(s) | Neonatal LOS and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | Study data derived from abstract only. Participant demographics or study period data not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Limited information from abstract. Controls not included in the analysis. | ||
| Patient characteristics and setting | Newborns with suspected sepsis admitted to the neonatal ward or NICU. Participant demographics or study period data not available. | ||
| Index tests | Real‐time PCR targeting 16S rRNA. | ||
| Target condition and reference standard(s) | Neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples were tested for routine culture and PCR separately. There was no mention if blood sample was drawn simultaneously. | ||
| Comparative | |||
| Notes | Abstract only. Participant demographics or study period data not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Neonates who met inclusion criteria during the study period were enrolled. Controls were not included in the analysis. | ||
| Patient characteristics and setting | Neonates aged 1 to 28 days with suspected sepsis admitted to the neonatal ward and NICU of Zhejiang University Children's Hospital from January 2005 to January 2007. 108 of the participants were preterm infants. | ||
| Index tests | Real‐time PCR with Gram‐specific probes followed by sequencing. | ||
| Target condition and reference standard(s) | Suspected neonatal EOS and LOS and blood culture. | ||
| Flow and timing | PCR and culture were done simultaneously. Unclear if samples were concurrently. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Infants were enrolled if they met inclusion criteria. | ||
| Patient characteristics and setting | Infants < 7 days old with suspected sepsis admitted to a level II NICU. Study period details not available. | ||
| Index tests | Broad‐range 16S rRNA PCR. | ||
| Target condition and reference standard(s) | Suspected neonatal sepsis and blood culture. | ||
| Flow and timing | Blood samples for PCR and culture were drawn concurrently. | ||
| Comparative | |||
| Notes | Study period details not available. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
CBC: complete blood count; CSF: cerebrospinal fluid; EDTA: ethylenediaminetetraacetic acid; EOS: early‐onset sepsis; LOS: late‐onset sepsis; n: number of participants; NICU: neonatal intensive care unit; NPV: negative predictive value; PCR: polymerase chain reaction; PICU: pediatric intensive care unit; PPV: positive predictive value; rDNA: ribosomal DNA; rRNA: ribosomal ribonucleic acid; RT‐PCR: real‐time polymerase chain reaction; SD: standard deviation; VLBW: very low birth weight.
Characteristics of excluded studies [ordered by study ID]
Ir a:
| Study | Reason for exclusion |
| All samples (CSF) were positive by culture for bacterial meningitis and not in the context of suspected infection. | |
| Urine instead of blood sample was used for broad‐range 16S rDNA in detecting neonatal septicemia. | |
| All samples investigated were culture negative samples and not in the context of suspected infection. | |
| GBS fluorescent PCR not compared with the reference standard (all were culture negative samples). | |
| Analyzed gastric aspirates by molecular methods for DNA load followed by sequencing and cultures. Neonates were suspected of sepsis but no details of blood cultures to diagnose sepsis were available. | |
| Culture‐positive specimens were examined for 16srRNA for PCR and sequencing. Not evaluated in the clinical context of suspected sepsis. | |
| Pyrosequencing used to identify bacteria from positive blood culture bottles. Not evaluated in the clinical context of suspected sepsis. | |
| It is unclear how many participants included in the study were neonates. Attempt made to contact author for details. | |
| Term neonates had risk factors of sepsis (maternal fever, unknown maternal GBS) but not suspected of having sepsis. Both blood cultures and PCR were negative in this cohort. | |
| Culture‐positive specimens and healthy controls were evaluated and not in the clinical context of suspected sepsis. | |
| No clinical specimens from neonates with suspected sepsis. Spiked samples were used. | |
| Non‐neonatal population. |
CSF: cerebrospinal fluid; GBS: group B streptococcus; PCR: polymerase chain reaction.
Data
Presented below are all the data for all of the tests entered into the review.
| Test | No. of studies | No. of participants |
| 1 All molecular tests Show forest plot | 35 | 7339 |
| Test 1  All molecular tests. | ||
| 2 Molecular tests: blood samples only Show forest plot | 32 | 6999 |
| Test 2  Molecular tests: blood samples only. | ||
| 3 Molecular tests with good methodologic quality Show forest plot | 22 | 4150 |
| Test 3  Molecular tests with good methodologic quality. | ||

Study flow diagram.

Deeks' funnel plot for publication bias.

Risk of bias and applicability concerns graph: review authors' judgments about each domain presented as percentages across included studies.

Risk of bias and applicability concerns summary: review authors' judgments about each domain for each included study.

Forest plot of 1 All molecular tests. CI: confidence interval; FN: false negative; FP: false positive; TN: true negative; TP: true positive.

Summary receiver operating characteristic plot of all molecular tests.
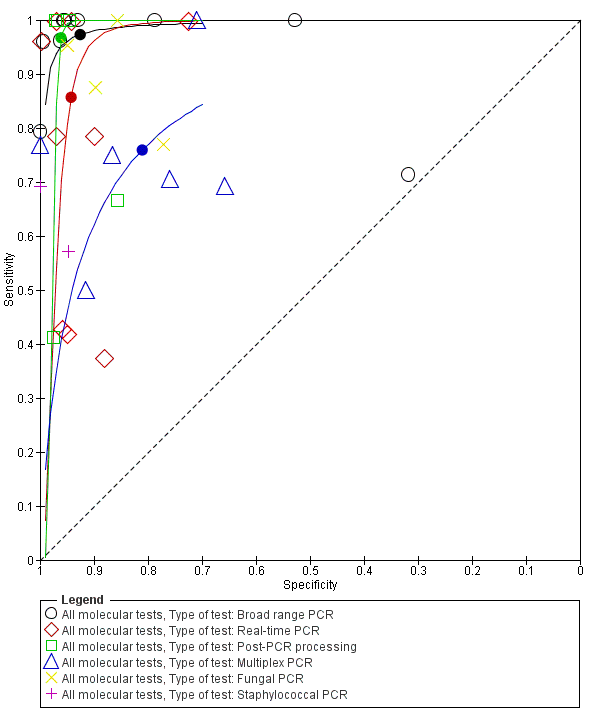
Summary receiver operating characteristic plot by type of molecular test. PCR: polymerase chain reaction.

Summary receiver operating characteristic plot subgrouped by sepsis onset.

Summary receiver operating characteristic plot subgrouped by gestational age.

Forest plot of all molecular tests sorted in order of prevalence. CI: confidence interval; FN: false negative; FP: false positive; TN: true negative; TP: true positive.

Summary receiver operating characteristic plot of all molecular tests where the size of the study symbol is directly proportional to the prevalence of sepsis in the study.
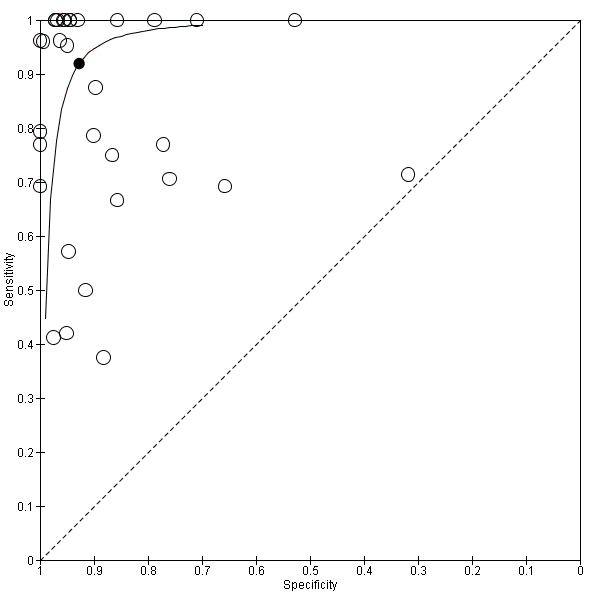
Summary receiver operating characteristic plot of studies that performed molecular tests on blood samples only.

Summary receiver operating characteristic plot of molecular tests with good methodologic quality.

All molecular tests.

Molecular tests: blood samples only.

Molecular tests with good methodologic quality.
| Groups | Number of studies | Sensitivity (95% CI) | Specificity (95% CI) | Quality of evidence using GRADE | |
| All studies | ‐ | 35 | 0.90 (0.82 to 0.95) | 0.93 (0.89 to 0.96) | Moderate quality evidence* |
| Type of test | Broad‐range PCR | 9 | 0.97 (0.86 to 1.00) | 0.93 (0.77 to 0.98) | Moderate quality evidence* |
| Real‐time PCR | 9 | 0.86 (0.59 to 0.96) | 0.94 (0.90 to 0.97) | Moderate quality evidence* | |
| Post‐PCR processing | 5 | 0.97 (0.40 to 1.00) | 0.96 (0.93 to 0.98) | Low quality evidence** | |
| Multiplex PCR | 6 | 0.76 (0.60 to 0.88) | 0.81 (0.70 to 0.89) | Low quality evidence** | |
| Staphylococcal PCR* | 2 | ‐ | ‐ | Low quality evidence** | |
| Fungal PCR* | 4 | ‐ | ‐ | Low quality evidence** | |
| Type of sepsis | EOS* | 2 | ‐ | ‐ | Low quality evidence** |
| LOS | 10 | 0.79 (0.69 to 0.86) | 0.94 (0.85 to 0.98) | Low quality evidence** | |
| Mixed EOS and LOS | 23 | 0.94 (0.84 to 0.98) | 0.92 (0.87 to 0.95) | Moderate quality evidence* | |
| Gestational age | Preterm | 5 | 0.89 (0.75 to 0.96) | 0.87 (0.71 to 0.94) | Low quality evidence** |
| Mixed term and preterm | 30 | 0.90 (0.80 to 0.96) | 0.94 (0.90 to 0.96) | Moderate quality evidence* | |
| Prevalence | < 15% | 20 | 0.94 (0.80 to 0.99) | 0.95 (0.92 to 0.97) | Moderate quality evidence* |
| 15% to 30% | 8 | 0.85 (0.67 to 0.94) | 0.88 (0.79 to 0.94) | Low quality evidence** | |
| >30% | 7 | 0.87 (0.75 to 0.93) | 0.93 (0.64 to 0.99) | Low quality evidence** | |
| Specimen | Blood only | 32 | 0.92 (0.84 to 0.96) | 0.93 (0.89 to 0.95) | Low quality evidence** |
| Blood and CSF* | 3 | ‐ | ‐ | Moderate quality evidence* | |
| Quality | Good methodologic studies only | 22 | 0.90 (0.78 to 0.96) | 0.93 (0.88 to 0.96) | Moderate quality evidence* |
| CI: confidence interval; CSF: cerebrospinal fluid; EOS: early‐onset sepsis; LOS: late‐onset sepsis; PCR: polymerase chain reaction. Summary estimates of sensitivity and specificity were derived from meta‐analyses using the bivariate random‐effects model using statistical software STATA. Summary estimates for the subgroups are presented, where number of studies ≥ 4. *Summary estimates of sensitivity and specificity could not be calculated using STATA if number of studies ≤ 4. GRADE rating of evidence: reasons for downgrading quality of evidence (Gopalakrishna 2014) * Evidence downgraded one level for inconsistency of evidence. ** Evidence downgraded two levels for inconsistency and imprecision. | |||||
| Test | No. of studies | No. of participants |
| 1 All molecular tests Show forest plot | 35 | 7339 |
| 2 Molecular tests: blood samples only Show forest plot | 32 | 6999 |
| 3 Molecular tests with good methodologic quality Show forest plot | 22 | 4150 |

