Pruebas genómicas prenatales no invasivas para la detección de la aneuploidía cromosómica fetal en embarazadas
References
References to studies included in this review
References to studies excluded from this review
References to ongoing studies
Additional references
References to other published versions of this review
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Jump to:
| Study characteristics | |||
| Patient sampling | Study design: case‐control study (1:2) from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 976 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 without multiplexing. Each library was sequenced using 50 bases‐length reads chemistry in a single end‐flow cell. Mean fetal fraction DNA: (male only) euploid: 20.11% and T21: 16.86%. In‐house gNIPT. | ||
| Target condition and reference standard(s) | Target condition: T21. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 50/275 samples were excluded during DNA extraction (47 for low amount of DNA and 3 for haemolysis) (no gNIPT results). 31/225 samples were excluded from analysis (8 for pretesting phase and 23 for reference set). 11/194 samples were excluded from analysis for insufficient fetal fraction DNA (no gNIPT results). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the implementation of gNIPT for trisomy 21 into a cytogenetics laboratory in a university teaching hospital as well as validate gNIPT’s clinical use on samples collected prospectively. | ||
| Funding source or sponsor of the study | Study not funded by industry. | ||
| Informations about the authors contacted | Authors were contacted on: 23 March and 4 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control study (1:3) from a prospective cohort. Inclusion criteria: singleton pregnancies between 11 to 13 weeks' gestation. | ||
| Patient characteristics and setting | Number enrolled: 400 pregnant women. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 25 samples did not meet Ariosa Diagnostics, Inc acceptance criteria but they were replaced with the next available cases. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess the prenatal detection rate of T21 and T18 and the false‐positive rate by chromosome‐selective sequencing of maternal plasma ccfDNA. | ||
| Funding source or sponsor of the study | Study not funded by industry but samples were analysed at Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded prospective cohort (second phase). First phase (case‐control study) not shown in the present review. | ||
| Patient characteristics and setting | Number enrolled: 2167 pregnant women. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Median fetal fraction DNA (range): euploids: 10.0% (4.1% to 31.0%) and T21: 14.0% (6.1% to 24.0%). | ||
| Target condition and reference standard(s) | Target condition: T13. | ||
| Flow and timing | Blood samples were obtained at the time of screening for euploid pregnancies (before reference standard). Blood samples were obtained after T13 confirmation following invasive procedure (reference standard). gNIPT was a first‐ or a second‐tier test. 165/2167 samples were excluded because they were used in the first phase. 53/2002 samples failed during amplification or sequencing (no gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess the performance of chromosome‐selective sequencing of maternal plasma cell‐free DNA (cfDNA) in non‐invasive prenatal testing for trisomy 13. | ||
| Funding source or sponsor of the study | Study not funded by industry but samples were analysed at Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, retrospective analysis from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 900 pregnant women. | ||
| Index tests | gNIPT by MPSS with Illumina v3 flow‐cell on a HiSeq 1500 sequencer in 12‐plex. Mean fetal fraction DNA: group 1 (patients without abnormal fetal ultrasound findings, but at high risk of fetal aneuploidy): 10.9% and group 2 (high risk of fetal aneuploidy after ultrasound finding): 11.2%. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 42 samples failed the initial MPSS testing for technical issues. 42/42 repeated tests using a second aliquot and 36/42 samples obtained gNIPT results. 6/892 samples failed during gNIPT process (low fetal fraction DNA or result appeared atypical) (no gNIPT result). | ||
| Comparative | |||
| Aim to study | To evaluate the performance of the gNIPT (using fetal ccfDNA) for detection of the 3 main autosomal fetal trisomies in a very high‐risk population of patients whose fetuses display ultrasonographically identified anomalies by comparing the results with those obtained by conventional fetal karyotyping. | ||
| Funding source or sponsor of the study | Funding source not reported. 1 author is an employee of Laboratoire CERBA and also a shareholder. | ||
| Informations about the authors contacted | Authors were contacted on: 25 May 2016. | ||
| Notes | Authors are from de Collaborative SEquençage a Haut Debit et Aneuploidies (SEHDA) Study Group. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 2362 pregnant women including 1847 singleton pregnancies (data not reported in the present review) and 515 twin pregnancies. Chorionicity (368/515): 13% monochorionic and 58.4% dichorionic. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Mean fetal fraction DNA (range): twins: 8.7% (4.1% to 30.0%) and singleton: 11.7% (4.0% to 38.9%). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 164/515 samples without follow‐up were excluded including138 lost to follow‐up, 19 still pregnant and 7 miscarriages or stillbirths without karyotype of fetal tissue. 29/515 samples failed the initial TMPS testing. 16/515 samples failed during sequencing process (no gNIPT result). | ||
| Comparative | |||
| Aim to study | To report the clinical implementation of cfDNA analysis of maternal blood in screening for T21, T18 and T13 in a large series of twin pregnancies and examine variables that could influence the failure rate of the test. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc made blinded sequencing and analysis. | ||
| Informations about the authors contacted | Author was contacted on: 1 June and 27 September 2016. No replies received from the author. | ||
| Notes | gNIPT results from singleton pregnancies were not reported in the present review for incomplete 2 x 2 tables. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control (1:4) study from the MELISSA prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 2882 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 6‐plex. Fetal fraction DNA: amount measured but not reported. 1) for T21, T18, and T13: positive if NCV > 4 (aneuploidy suspected if NCV is between 2.5 and 4). 2) for 45,X: positive if NCV for Chrom. X < ‐4 and NCV for Chrom. Y < 2.5. 3) for 47,XXX: positive if NCV for Chrom. X > 4 and NCV for Chrom. Y < 2.5. 4) for 47,XXY: positive if NCV for Chrom. X between ‐2.5 and 2.5 and NCV for Chrom. Y > 33. 5) for 47,XYY: positive if NCV for Chrom. X < ‐4 and NCV for Chrom. Y > 4 with NCV for Chrom. Y is 2 times greater than expected NCV Chrom. X. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXX, 47,XXY and 47,XYY. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 2091/2625 samples were not selected for this case‐control study. 2/534 samples were excluded for tracking issue. 16/532 samples without fetal DNA detected were excluded during process (no gNIPT result). 13/516 samples were excluded of T21 2 x 2 table for censored complex karyotype. 14/516 samples were excluded of T18 2 x 2 table for censored complex karyotype. 15/516 samples were excluded of T13 2 x 2 table for censored complex karyotype. 27/516 samples were excluded of 45,X 2 x 2 table for censored complex karyotype. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To prospectively determine the diagnostic accuracy of massively parallel sequencing to detect whole chromosome fetal aneuploidy from maternal plasma. | ||
| Funding source or sponsor of the study | Study funded by Verinata Health, Inc. (a wholly owned subsidiary of Illumina, Inc.). | ||
| Informations about the authors contacted | Authors were contacted on: 1 March and 30 November 2016. | ||
| Notes | This study is a clinical trial. MELISSA study. Clinicaltrials.gov NCT01122524. Data for 47,XXY, 47,XYY and 47,XXX were incomplete in the publication (data not shown in the present review). | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: retrospective study (archived maternal plasma samples) from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 2882 pregnant women. Median gestational age (range): 12.6 (10 to 21) weeks. | ||
| Index tests | gNIPT by MPSS with the sequencing chemistry Illumina TrueSeq 3.0. Fetal fraction DNA: not reported. 1) for T21, T18 and T13: positive if NCV > 4 (aneuploidy suspected zone between 3 and 4). 2) for 45,X: positive if NCV Chrom. X < ‐3 and NCV Chrom. Y < 3. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To estimate the accuracy and potential clinical effect of using massively parallel sequencing of maternal plasma DNA to detect fetal aneuploidy in a population of pregnant women carrying fetuses with nuchal cystic hygroma. | ||
| Funding source or sponsor of the study | Study funded by Verinata Health, Inc. (a wholly owned subsidiary of Illumina, Inc.). | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | 74/113 samples were previously sequenced during the MELISSA trial. In this study, all 113 samples were newly resequenced (no overlap) with MELISSA study. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 2052 pregnant women. Mean maternal age (± SD; range): 29.6 (± 5.54; 18 to 48.6) years. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 8‐plex. Mean fetal fraction DNA: more than 35 years old: 11.3%, less than 35 years old: 11.6%, and at third trimester only: 24.6%. The traditional screening tests (first‐trimester combined test or a second‐trimester result (quadruple, serum integrated, fully integrated or sequential)) were also assessed. Mixed cutpoints used. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior or after the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 72/2042 samples without clinical outcome. 38/2042 samples without standard screening result. 17/2042 samples without gNIPT result. 1/2042 samples without standard screening result and without gNIPT result. 12 resequenced samples were in the grey zone (between affected and unaffected) and were successfully resequenced in uniplex. | ||
| Comparative | |||
| Aim to study | To compare the results of gNIPT with ccfDNA for fetal autosomal aneuploidy with the results of conventional screening for T21 and T18 in a general obstetrical population. To compare false positive rates with the use of each method. To compare false positive rates for T13 in a subset of pregnant women in whom standard screening results included a risk assessment for trisomy 13. To compare fetal ccfDNA fractions in low‐risk patients and those in high‐risk patients in the CARE study population to assess the potential effects of demographic differences on test performance. | ||
| Funding source or sponsor of the study | Study funded by Illumina, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 10 February, 1 June and 28 June 2016. | ||
| Notes | This study is a clinical trial (Comparison of Aneuploidy Risk Evaluations; CARE study). ClinicalTrials.gov number: NCT0166335. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 10 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in multiplex with BGI's algorithm. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To present initial results of non‐invasive prenatal diagnosis of common aneuploidies (T21, T18, and T13) based on ccfDNA in maternal plasma in high‐risk pregnant women, and to compare the results with routine karyotyping. | ||
| Funding source or sponsor of the study | Study not funded by industry but NIFTY™ tests were provided by Beijing Genomics Institute, Shenzen, China. | ||
| Informations about the authors contacted | Authors were contacted on: 2 May and 4 July 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: case‐control study. | ||
| Patient characteristics and setting | Number enrolled: 4664 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 4‐plex. Fetal fraction DNA range: 7% to 55%. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T13. T18 was also assessed but no case was found. | ||
| Flow and timing | Blood samples were obtained immediately prior the invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample was reported in multifetal pregnancies. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To study prenatal testing for T21, T18, and T13 by MPSS of fetal ccfDNA in high‐risk multifetal pregnant women. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. Some authors are employees and shareholders of Sequenom, Inc. or of Sequenom Center for Molecular Medicine. | ||
| Informations about the authors contacted | Author was contacted on: 10 March 2016. Reply received on: 16 March 2016. | ||
| Notes | This study is a clinical trial "A New Prenatal Blood Test for Down Syndrome" ClinicalTrials.gov number: NCT00877292. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control study from a prospective cohort and archived plasma. Participants: pregnant women with clinical indications of fetal aneuploidy (high risk of fetal aneuploidy) for invasive procedure. Inclusion criteria: singleton pregnancies with and without trisomy 13, 18 or 21, matched for gestational ages. Exclusion criteria: twin pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 392 pregnant women (252 from the prospective cohort and 140 were archived plasma). Number available for 2 x 2 table: 289 pregnant women (subgroup of 74%). Setting: 10 centres in Hong Kong, the Netherlands, and UK. Recruitment period for the prospective cohort: October 2008 to May 2009. Recruitment period for the archived plasma samples collection: October 2003 to September 2008. Ethnicity: not reported. Gestational age: not reported. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx in 2‐plex. Feta fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3. Commercial test: Sequenom's test. | ||
| Target condition and reference standard(s) | Target conditions: T18 and T13. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 103/392 samples were selected as reference control. No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess the prenatal diagnostic performance by MPSS of maternal plasma DNA on a cohort of pregnant women with T13 and T18 fetuses. | ||
| Funding source or sponsor of the study | Study co‐sponsored by Sequenom, Inc and Life Technologies. Some authors have filed patent on gNIPT (part of this patent has been licensed to Sequenom, Inc). | ||
| Informations about the authors contacted | Author was contacted on: 14 December 2015 and 10 May 2016. Reply received on: 12 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:5) from a prospective cohort and archived plasma. Participants: pregnant women with clinical indications for invasive procedure, mixed risk (mostly high risk (> 1/300 at traditional screening test), intermediate risk (between 1/300 and 1/1000) or other risk factors). T21 and non T21 pregnancies matched for gestational ages. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 824 pregnant women. Number available for 2 x 2 table: 753 (8‐plex) (subgroup of 91%). Setting: 10 centres in Hong Kong, the Netherlands, and UK. Recruitment period for the prospective cohort: October 2008 to May 2009. Recruitment period for the archived plasma samples collection: October 2003 to September 2008. Ethnicity: not reported. Median gestational age: 13.1 weeks. Median maternal age: 35.4 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer II in 8‐plex and 2‐plex (not reported in the present review). Median fetal fraction DNA (interquartile 1 and 3): male euploid: 15.2% (10.6% and 19.1%), archived samples: 14.7%, and prospective samples: 15.4%. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3. Commercial test: Sequenom's test. | ||
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 60/824 samples were excluded before sequencing process (2 twin pregnancies, 12 without karyotype and 46 failed quality control for blood sampling). 11/764 samples failed quality control during sequencing process (no gNIPT result). 96/753 samples were also used for reference controls (8‐plex). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To validate the diagnostic performance and practical feasibility of massively parallel genomic sequencing for the non‐invasive prenatal assessment of trisomy 21 in pregnant women who had undergone conventional screening and were clinically indicated for definitive testing. | ||
| Funding source or sponsor of the study | Study sponsored by Sequenom, Inc. Some authors have filed patent applications on gNIPT (part of this patent has been licensed to Sequenom, Inc). | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | Data from 2‐plex sequencing were excluded from the present review to avoid double counting. We kept data from 8‐plex because it is the method most likely to be used for routine testing. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, observational prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 333 pregnant women (85.5% without prior risk and 16.5% were at high risk of fetal aneuploidy). | ||
| Index tests | gNIPT by TMPS (DANSR assay or SNP‐based method). Mean fetal fraction DNA (range): 12.7% (4.2% to 27.9%), Harmony™ prenatal test: 13.1%, and Panorama™ prenatal test: 12.7%. SNP‐based method cutpoint: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY. SCA data were not reported in the present review. T18 and T13 were also assessed but no case was found. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal clinical examination. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 17/333 samples excluded because still pregnant at the time of publication (no follow‐up). 9/333 samples failed the initial TMPS testing. 6/9 repeated sampling was performed and results were obtained in 5/6. 3/333 samples without gNIPT result were excluded (unrepeated samples). 1/333 samples without gNIPT result and follow‐up were excluded (still pregnant). | ||
| Comparative | |||
| Aim to study | To evaluate gNIPT of ccfDNA as a screening method for major chromosomal anomalies in a clinical setting. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc and Natera, Inc. made sequencing and analysis. | ||
| Informations about the authors contacted | Author was contacted on: 27 May 2016 and 31 May 2016. | ||
| Notes | gNIPT is offered to pregnant women at their own cost. 45,X, 47,XXY, 47,XYY and 47,XXX were also screened but inappropriate reference standard for the present review was used. gNIPT data from SCA were not shown in this review. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: retrospective cohort study. Data from prospective cohort were not shown in the present review. | ||
| Patient characteristics and setting | Number enrolled: 207 pregnant women from the retrospective cohort. Setting: 1 centre at Kings’ College Hospital in London, UK. Relevant tests carried out prior to index test: none. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Mean fetal fraction DNA (range): euploids: 9.8% (7.4% to 12.1%), T21: 10.8% (6.8% to 12.1%), and T13: 7%. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T13. T18 was also assessed but no case was found. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To examine the clinical implementation of TMPS of ccfDNA in maternal blood and an algorithm that relies on the lower fetal fraction DNA contribution of the 2 fetuses in the assessment of risk for trisomies in twin pregnancies. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc have performed gNIPT at their own expense. Study funded by a grant from The Fetal Medicine Foundation, UK. | ||
| Informations about the authors contacted | Author was contacted on: 27 May and 27 September 2016. No reply received from the author. | ||
| Notes | Data from prospective cohort study were not shown in the present review because patients with gNIPT negative result were without follow‐up to confirm gNIPT result. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:11) from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 480 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx in 4‐plex. Minimum fetal fraction DNA as estimated with the fetal quantifier assay: 3.9%. | ||
| Target condition and reference standard(s) | Target condition: T21. | ||
| Flow and timing | Blood samples were obtained prior or after the invasive procedure (reference standard). gNIPT was a second‐tier test. 13/480 samples excluded before sequencing process (9 for plasma volume < 3.5 mL and 4 for processing errors). 20/467 samples failed the initial MPSS testing. 18/467 samples failed quality control during sequencing process, including 7 samples for low fetal fraction DNA (no gNIPT result). | ||
| Comparative | |||
| Aim to study | To evaluate a multiplexed massively parallel shotgun sequencing assay for noninvasive trisomy 21 detection using circulating cell‐free fetal DNA. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | Author was been contacted on: 5 May and 28 September 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Retrospective cohort (training set) not reported in the present review. | ||
| Patient characteristics and setting | Number enrolled: 7103 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2500 sequencer in 15‐plex with SAFeR™ algorithm. Fetal fraction DNA: the limit of detection (the lowest fetal fraction DNA with a detectable aneuploidy) for T21 was determined at 2% fetal fraction level. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 79/100 repeated samples with a second blood draw and all obtained a gNIPT result. 21/100 unrepeated samples failed quality control metrics (no gNIPT result). | ||
| Comparative | |||
| Aim to study | To determine the limit of detection of a gNIPT method, in order to define the actual lower fetal fraction DNA required to detect common fetal autosomal trisomies, using a model system to simulate samples at different proportions of fetal ccfDNA. Secondly, to assess the impact of low fetal fraction DNA on the performance of ccfDNA‐based maternal plasma testing for aneuploidies. | ||
| Funding source or sponsor of the study | Study not funded by industry but the samples were analysed in the GENOMA laboratory (Rome, Italy). | ||
| Informations about the authors contacted | Authors were contacted on: 30 August and 6 September 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 11,692 pregnant women. Gestational age: not reported. | ||
| Index tests | gNIPT by TMPS (DANSR assay). Fetal fraction DNA: not reported. Traditional screening test was also assessed but 2 x 2 tables were incomplete. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 7994/11,692 samples did not undergo a gNIPT (no gNIPT result). 99/3698 samples failed the initial TMPS testing. 54/99 repeated sampling were processed and 34/54 gNIPT results were obtained. 65/3698 samples without gNIPT result. | ||
| Comparative | |||
| Aim to study | To report the feasibility of implementing gNIPT. To examine the factors affecting patient decisions concerning their options for screening and decisions on the management of affected pregnancies. To report the prenatal diagnosis of fetal trisomies and outcome of affected pregnancies following the introduction of contingent screening. | ||
| Funding source or sponsor of the study | Study not funded by industry but the cost of collection and analysis of the blood samples for the cell‐free DNA test was covered by Ariosa Diagnostics, Inc. These organisations had no role in study design, data collection, data analysis, data interpretation or writing of the report. Study was funded by a grant from The Fetal Medicine Foundation, UK. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| High | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: case‐control study (1:3), age‐matched randomly selected from a larger cohort. Inclusion criteria: pregnant women at least 18 years of age who had signed an informed consent, and with singleton pregnancy. | ||
| Patient characteristics and setting | Number enrolled: more than 1000 pregnant women. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencer. Samples were amplified using 11,000‐plex or 19,488‐plex targeted polymerase chain reaction (targets included SNPs from chromosomes 13, 18, 21, X, and Y). Mean fetal fraction DNA (median; range): 12.1% (11.1%; 2.2% to 30.4%). | ||
| Target condition and reference standard(s) | Target condition: T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. About 932 samples were not selected for this case‐control study. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To determine how a single nucleotide polymorphism (SNP)‐ and informatics‐based non‐invasive prenatal aneuploidy test performs in detecting trisomy 13. | ||
| Funding source or sponsor of the study | Study funded by Natera, Inc. (involved in study design, data collection and analysis, decision to publish, and preparation of the manuscript). | ||
| Informations about the authors contacted | Authors were contacted on: 21 April 2016, and 27 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: case‐control study from archived plasma samples from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: not reported. 432 maternal plasma samples were retrieved from the prospective cohort. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXY and 47,XXX. 47,XYY was also assessed but no case was found. | ||
| Flow and timing | Blood samples were obtained prior the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess the performance of a directed chromosomal analysis approach in the prenatal evaluation of fetal sex chromosome aneuploidy. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | BGI‐Shenzhen were contacted on: 19 May 2016. Author was contacted on: 16 June 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 308 pregnant women. | ||
| Index tests | gNIPT by MPSS on IIIumina HiSeq 2000 sequencer with BGI's algorithm. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, 45,X, 47,XXY and 47,XYY. T13 and 47,XXX were also assessed but no cases were found. | ||
| Flow and timing | Blood samples were obtained just prior the invasive procedure (reference standard). gNIPT was a second‐tier test. 103/308 patients did not undergo gNIPT (no gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate the clinical value of gNIPT using ccfDNA in maternal blood. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI‐Shenzhen provided the test. | ||
| Informations about the authors contacted | Author was contacted on: 11 April 2016 (author) and 19 May 2016 (BGI's contact). | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 189 pregnant women. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 platform. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess the performance of noninvasive prenatal testing for trisomies 21 and 18 on the basis of MPSS of ccfDNA from maternal plasma in twin pregnancies. | ||
| Funding source or sponsor of the study | Funded by the Shenzhen Engineering Laboratory for Clinical Molecular Diagnostic, the China National GeneBank‐Shenzhen, the Medical Centre for Critical Pregnant Women in Guangzhou and Prenatal monitoring, In utero therapy and Follow‐up after birth in the complexity of Twin Pregnancy. Some authors worked for BGI‐Shenzhen. | ||
| Informations about the authors contacted | Author was contacted on: 10 February 2016. BGI‐Shenzhen were contacted on: 19 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 1228 pregnant women screened at first‐trimester, including 1184 pregnant women with normal first‐trimester ultrasound and 44 with abnormal ultrasound. Recruitment period: June 2012 to January 2013. | ||
| Index tests | gNIPT by TMPS (DANSR assay). Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. | ||
| Comparative | |||
| Aim to study | To assess the performance of nuchal translucency measurement followed by gNIPT in the first‐trimester to screen for aneuploidy in a community‐based average‐risk population. | ||
| Funding source or sponsor of the study | Funding source not reported but 1 author is employed by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 22 February 2016 and 15 March 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 155 pregnant women. | ||
| Index tests | gNIPT by MPSS on Ion Torrent PGM sequencer with 10 samples per chip. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigated whether fetal T18 and T21 were sensitively and specifically detectable by semiconductor sequencer: Ion Proton™. | ||
| Funding source or sponsor of the study | Study funded by the Industrial Strategic Technology Development Program, "Bioinformatics platform development for next generation bioinformation analysis" funded by the Ministry of Knowledge Economy (MKE, Korea). | ||
| Informations about the authors contacted | Author was contacted on: 6 and 11 April 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| High | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 903 pregnant women. | ||
| Index tests | gNIPT by MPSS on platforms Illumina Genome Analyzer IIx or Illumina HiSeq 2000 by multiplex sequencing. Fetal fraction DNA (range): quality control criteria > 3.5% (1% to 33%). 1) Positive if binary hypothesis t score (first hypothesis) > 3 and t score (second hypothesis) < 3 and if logarithmic LR > 1 (autosomal aneuploidy). 2) Positive if t score < ‐2.5 (45,X and 47,XXX) without Chrom. Y representation. 3) Positive if t score > 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X and Y independently (47,XXY and 47,XYY) for male fetus. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. | ||
| Flow and timing | It is not reported if the blood samples were obtained prior or after the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To develop an advanced gNIPT method based on MPSS. | ||
| Funding source or sponsor of the study | Study funded by industry. BGI was involved in the study design, conduct of the study, analysis and interpretation of results. | ||
| Informations about the authors contacted | Author was contacted on: 19 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 375 pregnant women (184 for the validation set). | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer in 5‐plex. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior the invasive procedure (reference standard). 191/375 not selected, samples for the validation set were excluded. 2/173 samples were resequenced because gNIPT results were in the inconclusive zone and 2 results were obtained. | ||
| Comparative | |||
| Aim to study | To introduce gNIPT for fetal autosomal trisomies and gender in a Danish public health setting, using semi‐conductor sequencing and published open source scripts for analysis. | ||
| Funding source or sponsor of the study | No funding source was reported. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 2340 pregnant women. | ||
| Index tests | gNIPT by MPSS. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To validate the efficacy of detection of fetal cell‐free DNA in maternal plasma of trisomy 21, 18 and 13 in a clinical setting. | ||
| Funding source or sponsor of the study | Study not funded by industry but patients had obtained insurance plans on behalf of Shenzhen Huada Genomics Institute. | ||
| Informations about the authors contacted | Author was contacted on: 22 April 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 101 pregnant women. | ||
| Index tests | gNIPT by MPSS on Ion Torrent PGM (data not shown in the present review) and Ion Proton™ sequencer in multiplex. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target condition: T21. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To compare the Ion Torrent PGM and Ion Proton™ platforms for gNIPT for fetal T21 directly using PGM and Ion Proton™ simultaneously for the same set of samples. | ||
| Funding source or sponsor of the study | Study funded by Genome Care internal research funding. The first author is employee of Genome Care. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | Data from PGM sequencer are not shown in the present review to avoid patients overlap. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| High | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 1968 pregnant women. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers with NATUS algorithm. Fetal fraction DNA: not reported (usually NATERA used quality control criteria > 4%). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. 45,X, 47,XXY, 47,XYY and 47,XXX were also screened but inappropriate reference standard for the present review was used (data not shown in this review). | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 240/1968 samples did not undergo gNIPT (no gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To examine possibility to use combination of gNIPT and chromosomal microarray analysis for prenatal diagnostics and their advantages between combined first‐trimester screen with confirmation by karyotyping of CVS or amniocytes. | ||
| Funding source or sponsor of the study | Study not funded by industry but gNIPT was carried out by Natera, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 21 June 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women mostly at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: not reported. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 108 pregnant women. Number available for 2 x 2 table: 108 pregnant women (whole cohort included in analyses). Setting: 1 centre in Japan. Recruitment period: not reported. Ethnicity: Asian. Median gestational age (range): 12.7 (11.6 to 28) weeks, 89.8% < 14 weeks. Mean maternal age (± SD): 37 (± 4.3) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected immediately before reference standard. Cutpoint: 1) positive if Z score ≥ 3 (T21, T18 and T13). 2) for female fetus, positive if Chrom. X Z score ≤ ‐3 (45,X). 3) for female fetus, positive if Chrom. X Z score ≥ 3 (47,XXX). 4) for male fetus, positive if Chrom. Y Z score ≥ 3 (47,XXY). Commercial test: NIFTY™ prenatal test by BGI‐Shenzhen. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X and 47,XXY. 47,XYY and 47,XXX were also assessed but no case was found. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were collected immediately before invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate the inclusion of an internal reference in the noninvasive prenatal identification of common fetal aneuploidies using massively parallel sequencing on maternal plasma. | ||
| Funding source or sponsor of the study | Study funded by BGI‐Shenzhen. | ||
| Informations about the authors contacted | BGI‐Shenzhen contacted on: 19 May 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 93 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina MiSeq sequencer in 12‐plex or on NextSeq 500 sequencer in 96‐plex. Median fetal fraction DNA (range): male fetus only: 10.2% (3.85% to 25.0%). 1) positive if Z score > 4 (intermediate risk if Z score between 2.5 and 4) for T21 and T18. 2) positive if Z score > 2.8 (intermediate risk if Z score between 1.9 and 2.8) for T13. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. SCA were also assessed but no case was found. | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior to the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the performance of MomGuard™, a gNIPT, for detecting T21, T18, T13, and SCA abnormalities recently developed in Korea. | ||
| Funding source or sponsor of the study | Study funded by a grant from the LabGenomics Clinical Research Institute. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: Retrospective cohort, blinded case‐control study. | ||
| Patient characteristics and setting | Number enrolled: 5321 pregnant women in all 4 cohorts. 1222 pregnant women selected for this study. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 6‐plex or uniplex. Fetal fraction DNA: amount measured but not reported. 1) positive for T21 if Z score ≥ 3. 2) positive for T18 or T13 if Z score ≥ 3.95. 3) positive for 45,X if Z score < ‐3.5 (non‐reportable regions between ‐2.5 and ‐3.5). 4) positive for 47,XXX if Z score > 3.5 (non‐reportable regions between 2.5 and 3.5). 5) positive for 47,XYY if Z score < ‐3.5 with Chrom. Y representation. 6) positive for 47,XXY if Z score is between ‐3.5 and 3.5 with Chrom. Y representation. Commercial test: Sequenom's test. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. copy number variants ≥ 7 Mb were also assessed but data not shown in the present review. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior or after the invasive procedure (reference standard). 4099/5321 samples not selected for this study. 42/1208 samples failed during autosome sequencing process (no gNIPT result) including 11/42 failed samples for low fetal fraction DNA, 29/42 failed samples for technical reasons and 2/42 failed samples for other biological reasons (maternal event). 22/1166 samples failed SCA sequencing process (no gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To provide a clinical validation of the sensitivity and specificity of a novel NIPT for detection of genome wide abnormalities. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 435 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 8‐plex or 12‐plex. Fetal fraction DNA: for a Z score cutoff value of 3 for chromosome 21, fetal DNA was estimated to 5.52%. 1) positive if Z score > 3 (T21). 2) positive if Z score > 5.91 (T18). 3) positive if Z score > 5.72 (T13). 4) positive if Z score Chrom. X < ‐2.91 and Z score Chrom. Y < 3 (45,X). 5) positive if Z score Chrom. X range from ‐2.91 to +2.91 and Z score Chrom. Y > 3 (47,XXY). 6) positive if Z score Chrom. X > 2.91 and Z score Chrom. Y < 3 (47,XXX). 7) positive if Z score Chrom. X < ‐2.91 and Z score Chrom. Y > 3 (47,XYY). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY, and 47,XXX. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 11/423 samples without karyotype were excluded (no reference standard result). | ||
| Comparative | |||
| Aim to study | To determine whether gNIPT by maternal plasma DNA sequencing can uncover all fetal chromosome aneuploidies in 1 simple sequencing event. | ||
| Funding source or sponsor of the study | Study not funded by industry but Berry Genomics Co. Ltd performed the sequencing analysis for free. This study was supported by the grants from the National High Technology Research and Development Program of China (863 Program) (No.2011AA02A112), the National Key Basic Research Program of China (2012CB944600) and the National Key Technology R&D Program of China (2012BAI09B05). | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 153 pregnant women. Setting: Henan Province People Hospital Medical. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq sequencer in multiplex. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X and 47,XYY. 47,XXY and 47,XXX were also assess but no case were found. | ||
| Flow and timing | Blood samples for gNIPT were obtained 30 minutes prior to the invasive procedure (reference standard). No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To determine the feasibility and accuracy of detecting numerical chromosomal abnormalities by high‐flux sequencing analysis of ccfDNA from maternal plasma. | ||
| Funding source or sponsor of the study | Study funded by by the Nalional Natural Science Foundation of China and a Medical Science and Technology Research Project of Henan Province. | ||
| Informations about the authors contacted | Author was contacted on 11 April 2016 but contact author's email is no longer valid. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective and retrospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 10,598 pregnant women. 2439 from prospective cohort and 8159 from retrospective cohort. | ||
| Index tests | gNIPT by MPSS on BGISEQ‐1000 platform in 16 or 24‐plex. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐ or a first‐tier test. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To report the established gNIPT screening system and the clinical performance of a new ultrahigh throughout gNIPT method based on combinatorial probe‐anchor ligation sequencing (cPAL) of ccfDNA in detecting T21, T18 and T13 in the multicentre network using a centralised testing mode. | ||
| Funding source or sponsor of the study | Some authors are employees of BGI‐Shenzhen, BGI‐Manufacture or BGI‐DX. Study funded by Shenzhen Birth Defect Screening Project Lab, Key Laboratory of Cooperation Project in Guangdong Province, Shenzhen Municipal Government of China, Pilot projects of regional strategic emerging industry cluster development by Hubei provincial development and Reform Commission and Action plan for the development of high‐tech industry in biotechnology and new medicine in 2012 by Wuhan Science and Technology Bureau. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study (validation set). | ||
| Patient characteristics and setting | Number enrolled: 1975 pregnant women including 1564 in the training set (data not shown in the present review) and 411 in the validation set. | ||
| Index tests | gNIPT by MPSS on Illumina v3 flow cells on HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. 1) positive for 45,X if Z score < ‐3.5 (non‐reportable regions between ‐2.5 and ‐3.5). 2) positive for 47,XXX if Z score > 3.5 (non‐reportable regions between 2.5 and 3.5). 3) positive for 47,XYY if Z score < ‐3.5 with Chrom. Y representation. 4) positive for 47,XXY if Z score is between ‐3.5 and 3.5 with Chrom. Y representation. Commercial test: Sequenom's prenatal test. | ||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXY, 47,XYY and 47,XXX. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 1564/1975 excluded samples were used for the training set. 21/411 failed samples were in the non reportable region and were considered positive gNIPT result by authors. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To extend the detection of autosomal aneuploidies by MPSS of ccfDNA from maternal plasma to include common sex chromosome aneuploidies. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. and Sequenom Center for Molecular Medicine (SCMM). | ||
| Informations about the authors contacted | Author was contacted on: 26 May 2016. | ||
| Notes | Data from the training set were not shown in the present review. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: retrospective study from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 2230 pregnant women. | ||
| Index tests | gNIPT by TMPS (DANSR assay). Median fetal fraction DNA (interquartile range): euploids: 10.0% (7.8% to 13.0%), T21: 12.5% (9.2% to 21.3%), and T18: 9.3% (5.6% to 13.0%). The traditional screening test (combined test at the first trimester) was also assessed. Cutpoint of combined test: 1 in 150. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐tier test. 100/2049 samples failed during sequencing process including 46 for low fetal DNA and 54 had assay failures (no gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess performance of noninvasive prenatal testing for fetal trisomy in a routinely screened first‐trimester pregnancy population. | ||
| Funding source or sponsor of the study | The study was supported by a grant from the Fetal Medicine Foundation (UK). The cost of collection and analysis of the samples was covered by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 242 pregnant women. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers with NATUS algorithm. Fetal fraction DNA: the lowest fetal fraction DNA on a case that returned a result was 3.95%. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, 45,X. 47,XXY, 47,XYY and 47,XXX were also assessed but no case was found. T13 was also assessed but the only 1 case presented in this publication was published thereafter in Hall 2014. T13 case was excluded to avoid double counting. | ||
| Flow and timing | Blood samples were obtained just before the invasive procedure (reference standard). gNIPT was a second‐tier test. No repeated test reported. 1 T13 cases was excluded to avoid double counting because it was published thereafter in Hall 2014. | ||
| Comparative | |||
| Aim to study | To assess the performance of ccfDNA testing in maternal blood for detection of fetal aneuploidy of chromosomes 13, 18, 21, X, and Y using TMPS of single‐nucleotide polymorphisms. | ||
| Funding source or sponsor of the study | Study funded by a grant from the Fetal Medicine Foundation (UK Charity No: 1037116). Analysis of samples was performed at their own expense by Natera, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | T13 cases data are not shown in the present review. They were excluded to avoid double counting because they are also published in Hall 2014. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: case‐control study. Participants: pregnant women selected from a high‐risk population (archived maternal plasma samples). Inclusion criteria: singleton pregnancies. Exclusion criteria: cases of fetal mosaicism and multifetal pregnancies. | ||
| Patient characteristics and setting | Recruited participants: 177 archived maternal plasma. Number available for 2 x 2 table: 172 samples (subgroup of 97%). Setting: recruitment in London, UK. Ethnicity: Caucasian (90%), Afro‐Caribbean (4%), Asian (5%) and other (1%). Gestational age range: 11.2 to 14.1 weeks. Maternal age range: 17.3 to 47.8 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Median fetal fraction DNA (range): euploids: 13.0% (4.8% to 32.0%), 45,X: 10.0% (6.3% to 18.0%), and 47,XXX, 47,XXY, and 47,XYY: 12.0% (6.4% to 16.0%). Blood samples for gNIPT were collected just before reference standard. Cutpoint: positive if FORTE algorithm risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. | ||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXX, 47,XXY, and 47,XYY. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were collected just before invasive procedure (reference standard). gNIPT was a second‐tier test. 5/177 samples failed during sequencing process (no gNIPT result), including 1 sample failed laboratory quality control metrics and 4 samples failed for an insufficient fetal ccfDNA fraction. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To report the clinical performance of chromosome‐selective sequencing of cfDNA in maternal blood and the FORTE algorithm for the assessment of fetal sex chromosome aneuploidies. | ||
| Funding source or sponsor of the study | No funding source was reported. | ||
| Informations about the authors contacted | Author was contacted on: 10 February 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 4002 pregnant women. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Mean fetal fraction DNA (± SD; range): euploids: 11% (± 4.5%; 4.2% to 51.3%), T21: 11.6% (± 4.2%; 5.1% to 23.3%), and T18: 10% (± 3.8%; 4.9% to 20.8%). | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 148/3228 samples failed during sequencing process (no gNIPT result), including 57 samples failed for low fetal fraction DNA and 91 samples failed sequencing process. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate performance of a gNIPT of fetal T21 and T18. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: unselected population of pregnant women undergoing aneuploidy screening (without prior risk of fetal aneuploidy). Inclusion criteria: singleton pregnancies, at least 18 years of age, and between 10 to 14 weeks of gestation. Exclusion criteria: women who had a miscarriage, chose to terminate the pregnancy or had a stillbirth without confirmatory genetic testing. | ||
| Patient characteristics and setting | Number enrolled: 18,955 pregnant women. Number available for 2 x 2 table: 15,841 pregnant women (subgroup of 84%). Setting: 35 centres in USA States, Canada, Sweden, the Netherlands, Belgium, and Italy. Recruitment period: March 2012 to April 2013. Ethnicity: Caucasian (70.9%), Afro‐American (8.2%), Asian (10.5%), Native American (0.6%), multiracial (2.7%), other (6.7%) and missing data (0.5%). Mean gestational age (range): 12.5 (10.0 to 14.3) weeks. Mean maternal age (range): 31 (18 to 48) years whose 76% of pregnant women analysed had < 35 years old. Relevant tests carried out prior to index test: none. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 sequencer in 96‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. The traditional screening tests (combined test at the first trimester) were also assessed. Cutpoint of combined test: 1 in 270 for T21 or 1 in 150 for T18 and T13. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi, amniotic fluid or products of conception or neonatal karyotype, neonatal clinical examination or medical record from birth. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐tier test. 3114/18,955 samples excluded of analysis including 229 samples did not meet inclusion criteria or meet exclusion criteria, 31 had twins, 121 had unknown ovum‐donor status, 64 withdrew or were withdrawn by investigator, 384 had sample‐handling errors, 308 without standard screening test result, 488 failed sequencing and have no gNIPT result (192 for low fetal fraction DNA, 83 for non fetal fraction DNA and 213 for high assay variance or assay failures) and 1489 were lost to follow‐up. | ||
| Comparative | |||
| Aim to study | To test the hypothesis that gNIPT has better performance than standard first‐trimester screening (with measurement of nuchal translucency and biochemical analytes) in risk assessment for trisomy 21 in a large, unselected population of women presenting for aneuploidy screening. To also evaluate the performance of gNIPT and standard screening in the assessment of risk for trisomies 18 and 13. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc and Perinatal Quality Foundation. | ||
| Informations about the authors contacted | Author was contacted on: 10 February 2016. Reply received on: 11 February 2016. | ||
| Notes | This study is a clinical trial (Noninvasive Examination of Trisomy (NEXT) ClinicalTrials.gov number, NCT01511458). | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control (1:3) study. | ||
| Patient characteristics and setting | Number enrolled: 4664 pregnant women. 1776 pregnant women selected for this study and 212 reanalysed samples from Palomaki 2011. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 4‐plex. Mean (geometric) fetal fraction DNA (range): 13.4% (4% to 50%). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained immediately prior the invasive procedure (reference standard). gNIPT was a second‐tier test. 110/1776 samples failed the initial MPSS testing. 105/110 samples required repeat testing using a second aliquot and 5/110 samples were resequenced with the same library. 93/110 samples obtained a gNIPT results. 17/1776 samples failed during sequencing process, most for low fetal fraction DNA (no gNIPT result). | ||
| Comparative | |||
| Aim to study | To determine whether maternal plasma ccfDNA sequencing can identify T18 and T13 as well as T21. | ||
| Funding source or sponsor of the study | Study fully funded by Sequenom, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | This study is a clinical trial "A New Prenatal Blood Test for Down Syndrome" ClinicalTrials.gov number: NCT00877292. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:9). | ||
| Patient characteristics and setting | Number enrolled: 442 pregnant women. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer in 8‐plex. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 11/437 twin pregnancies were not selected. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate the accuracy of the IONA® test in the discrimination between euploid pregnancies and those affected by fetal trisomies 21, 18 and 13. | ||
| Funding source or sponsor of the study | Study funded by Premaitha Health (public limited company). Some authors are employees of Premaitha Health plc. | ||
| Informations about the authors contacted | Author was contacted on: 19 September 2016. | ||
| Notes | Data from singleton pregnancies only reported here. See Papageorghiou 2016b for data on twin pregnancies. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:9). | ||
| Patient characteristics and setting | Number enrolled: 442 pregnant women. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer in 8‐plex. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 5/442 samples failed during sequencing process including 3 samples for low fetal fraction DNA and 2 samples did not have sufficient DNA fragment counts (no gNIPT result). 426/437 singleton pregnancies were not selected. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate the accuracy of the IONA® test in the discrimination between euploid pregnancies and those affected by fetal trisomies 21, 18 and 13. | ||
| Funding source or sponsor of the study | Study funded by Premaitha Health (public limited company). Some authors are employees of Premaitha Health plc. | ||
| Informations about the authors contacted | Author was contacted on: 19 September 2016. | ||
| Notes | Data from twin pregnancies only reported here. Data from singleton pregnancies reported in Papageorghiou 2016a. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 1064 pregnant women. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers, 19,488‐plex targeted PCR with NATUS algorithm. Range fetal fraction DNA: 2% to 50%. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior (93%) or after (7%) to the invasive procedure (reference standard). 13/1064 samples excluded for other aneuploidies, including 6 cases with triploidy, 3 fetal mosaics, 2 cases with 47,XXY, 1 case with 47,XXX and 1 case with 47,XYY. 85/1051 samples failed quality control (no gNIPT result) including 64 low fetal fraction DNA, 12 low DNA, 6 contaminations, 2 loss of heterozygosity and 1 poor model fit. Between 1 to 3 samples did not passed quality control for all 5 chromosomes. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To estimate performance of a single nucleotide polymorphism–based gNIPT (TMPS) for fetal aneuploidy in high‐risk and low‐risk populations on single venipuncture. | ||
| Funding source or sponsor of the study | Study funded by Natera, Inc. and a grant from the National Institute of Health, National Institute of Child Health and Human Development (4R44HD062114‐02). The majority of the authors are employees of Natera, Inc. and hold stock or options to hold stock in the company. | ||
| Informations about the authors contacted | Author was contacted on: 22 June 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 259 pregnant women. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers, 19,488‐plex targeted PCR with NATUS algorithm. Fetal fraction DNA: amount measured but not reported (usually NATERA used quality control criteria > 4%). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY and 47,XXX. 47,XYY was also assessed but no case was found. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate a strategy for clinical implementation of ccfDNA testing in high‐risk pregnancies after first‐trimester combined screening. | ||
| Funding source or sponsor of the study | Study not funded by industry but the cost of ccfDNA testing were covered by Natera, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: retrospective cohort, blinded nested case‐control study. | ||
| Patient characteristics and setting | Number enrolled: 242 pregnant women. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer. Fetal fraction DNA: amount measured but not reported. Commercial test: IONA® test by Premaitha Health (public limited company). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained just before the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To assess the potential performance of screening for fetal T21, T18 and T13 by ccfDNA analysis of maternal blood using the IONA® test. | ||
| Funding source or sponsor of the study | Study not funded by industry but the IONA® test was provided by Premaitha Health plc, Manchester, UK. Study supported by a grant from The Fetal Medicine Foundation. | ||
| Informations about the authors contacted | Author was contacted on: 19 September 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort, observational study. | ||
| Patient characteristics and setting | Number enrolled: 4170 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 12‐plex. Range fetal fraction DNA: 4% to 50%. 1) for T21, positive if Z score ≥ 3. 2) for T18 and T13, positive if Z score ≥ 3.95. 3) positive for 45,X if Z score < ‐3.5 (non‐reportable regions between ‐2.5 and ‐3.5). 4) positive for 47,XXX if Z score > 3.5 (non‐reportable regions between 2.5 and 3.5). 5) positive for 47,XYY if Z score risk < ‐3.5 with Chrom. Y representation. 6) positive for 47,XXY if Z score risk is between ‐3.5 and 3.5 with Chrom. Y representation. Commercial test: Sequenom's prenatal test. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXX, 47,XXY and 47,XYY. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. For autosomes: 54/3430 autosomes samples excluded for quality control deviation (low fetal DNA fraction, library concentration, total counts, and amplification bias). For autosomes: 54/3376 samples excluded for complex autosome karyotypes (mosaic, triploidies, unbalanced rearrangements with missing or duplicated genetic material). For 45,X and 47,XXX: 102/3430 samples excluded for low fetal fraction DNA or copy number variation of the Chrom. X is confounded by maternal component and cannot be determined. For 45,X and 47,XXX: 50/3328 samples excluded for complex SCA karyotype. For 47,XXY and 47,XYY: 182/3430 samples excluded for low fetal fraction DNA or copy number variation of the Chrom. X is confounded by maternal component and cannot be determined. For 47,XXY and 47,XYY: 47/3248 samples excluded for complex SCA karyotype. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To validate the clinical performance of MPSS of ccfDNA contained in specimens from pregnant women at high risk of fetal aneuploidy to test fetuses for T21, T18, T13, 45,X, 47,XXX, 47,XXY and 47,XYY. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 30 May 2016. | ||
| Notes | This study is a clinical trial (Non‐Invasive Screening for Fetal Aneuploidy) ClinicalTrials.gov number, NCT00847990. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 2905 pregnant women. | ||
| Index tests | gNIPT by TMPS (DANSR assay). Median fetal fraction DNA (range): 11% (4% to 40%). The traditional screening tests (combined test at the first trimester) was also assessed. Cutpoint of combined test: 1 in 100 for T21. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to invasive procedure (reference standard). gNIPT was a first‐tier test. 122/2905 failed the initial TMPS testing (122 = 123 ‐ 1 sample lost in mail). 66/2851 samples without follow‐up were excluded. 110/122 required repeat testing using a second blood sample and results were obtained in 69/110 samples. 53/2905 samples failed during sequencing process (41 samples failed second sequencing and 12 unrepeated tests) (no gNIPT result). | ||
| Comparative | |||
| Aim to study | To examine, in a general population (pregnant women without prior risk of fetal aneuploidy), the performance of ccfDNA testing for T21, T18 and T13 at 10 to 11 weeks’ gestation and compare it to that of the combined test at 11 to 13 weeks' gestation. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc made sequencing and analyses. | ||
| Informations about the authors contacted | Author was contacted on: 21 April 2016 and 30 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 201 pregnant women. Gestational age range: overall 9.4 to 36.4 weeks. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina HiSeq 2000 sequencer with NATUS algorithm. Mean fetal fraction DNA: euploids: 10.9% and aneuploids: 12.1%. Overall range: 2.9% to 37.7%. | ||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXX, 47,XXY, and 47,XYY. | ||
| Flow and timing | Blood samples were collected just before or at least 4 days after invasive procedure (reference standard). 1/187 sample excluded for conflicting algorithm metrics (no meaningful gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To develop a SNP‐based and informatics‐based gNIPT that detects sex chromosome aneuploidies early in pregnancy. | ||
| Funding source or sponsor of the study | It is unclear if the study was funded by industry but all authors are employees of Natera, Inc. except the first author (Carole Samango‐Sprouse). This study was supported in part by a grant from the National Institute of Health, National Institute of Child Health and Human Development. | ||
| Informations about the authors contacted | Author was contacted on: 22 April, 4 July and 29 September 2016. Replies received on: 29 and 30 September 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded retrospective study (archived maternal plasma samples). | ||
| Patient characteristics and setting | Number enrolled: overall: 1014 pregnant women including 71 women selected on 435 for the training set (not shown in the present review) and 48 women selected on 575 for the test set. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx sequencer in uniplex. Fetal fraction DNA: not reported. 1) positive if NCV > 4 for autosomes. There is a "no call zone" between 2.5 and 4 considering as gNIPT positive result for the present review. 2) positive if NCV for Chrom. Y < ‐2.0 SDs from the mean of male samples and if NCV for Chrom. X < ‐3.0 SDs from the mean of female samples for 45,X. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 895/1014 samples were not selected for sequencing. 71/119 samples were selected for the training set (not shown in the present review). 1/48 sample from twin gestation in the test set was removed from the final analysis. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To develop and test an optimised algorithm from MPSS data and demonstrated the potential universality of the sequence tag mapping and chromosome quantification method for the detection of multiple chromosomal abnormalities. | ||
| Funding source or sponsor of the study | Study funded by Illumina (formerly Verinata Health). The funding organizations played a direct role in the design of the study, the choice of enrolled patients, the review and interpretation of data, and the preparation and final approval of the manuscript. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 201 pregnant women. Gestional age range: overall 12 to 20 weeks. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. 1) positive if Z score > 3 (T21, T18, and T13). 2) positive if Z score Chrom. X < ‐3 and Z score Chrom. Y < 3 (45,X). 3) positive if Z score Chrom. X < ‐3 and Z score Chrom. Y > 3 (47,XYY). The traditional screening test (combined test at the first trimester) was also assessed but complete data for 2 x 2 tables were unavailable. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, and 47,XYY. 47,XXX and 47,XXY were also screened but no case was found. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the performance of gNIPT for all fetal chromosomal aneuploidies in an extremely high‐risk group undergoing first‐trimester combined T21 screening. | ||
| Funding source or sponsor of the study | Funding sources were not reported but 2 authors are affiliated to Berry Genomics Co. Ltd., Beijing, PR China. | ||
| Informations about the authors contacted | Author was contacted on: 10 February and 23 June 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 1916 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. The traditional screening test (second‐trimester triple test) was also assessed. Cutpoint of triple test: 1 in 270 for T21 and T18. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. 45,X, 47,XXX, 47, XXY, 47,XYY were also screened but inappropriate reference standard for the present review was used. | ||
| Flow and timing | It is not reported if the blood samples were collected before or after invasive procedure (reference standard). 102/1916 samples without follow‐up were excluded. 9/1916 samples were without follow‐up and failed sequencing process (no gNIPT result). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the performance of gNIPT for detection of fetal aneuploidies in a Chinese cohort of women younger than 35 years old in a prospective clinical setting. Also, to compare the performance of gNIPT with the routine prenatal screening (second‐trimester combined test). | ||
| Funding source or sponsor of the study | Study not funded by industry. This study was supported by a grant (2006BAI05A10) from the National Key Technology Research and Development Program of China during the ‘11th Five‐Year Plan’. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | SCA were also screened but inappropriate reference standard for the present review was used. gNIPT data from SCA were not shown in this review. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 213 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 sequencer in 12‐plex. Median fetal fraction DNA (range): only male fetus: 8.54% (2.69% to 18.75%). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY and 47,XXX. 47,XYY were also assessed but no case was found. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior the invasive procedure (reference standard). 8/212 samples without reference standard were excluded including 5 miscarriages, 2 intrauterine fetal deaths and 1 termination of pregnancy. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the feasibility of gNIPT of maternal plasma samples collected from pregnant Chinese women in early gestation, between 8 and 12.9 weeks’ gestation. | ||
| Funding source or sponsor of the study | Study not funded by industry but gNIPTs were done and analysed at Berry Genomics Co. Ltd. Study funded by a grant from the National Natural Science Foundation of China. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: case‐control study from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: not reported. A subset of 338 pregnant women including 171 women in the training set (data not shown in the present review) and 167 women in the validation set were selected for this study. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 sequencer in multiplex with FORTE algorithm. Range fetal fraction DNA: 3% to 33%. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. | ||
| Flow and timing | It is not reported if the blood samples were collected before or after invasive procedure (reference standard). No failed sample reported in the validation set. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To develop a novel biochemical assay and algorithm for the prenatal evaluation of risk for fetal T21 and T18 using ccfDNA obtained from maternal blood. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. All authors are employees of Aria Dx Inc. (now Ariosa Diagnostics). K Sparks is a member of the board of the company. | ||
| Informations about the authors contacted | Author was contacted on: 23 June 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Blinded for T21 and unblinded for T18 and T13. | ||
| Patient characteristics and setting | Number enrolled: 522 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 12‐plex with DAP.21 algorithm without CG correction. Mean fetal fraction DNA (range): male fetus only: 12.3% (3.7% to 36.8%). 1) positive if MAD‐based Z ‐score ≥ 3 for T21. 2) positive if MAD‐based Z score ≥ 3.2 for T18. 3) positive if MAD‐based Z score ≥ 3.9 for T13. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior the invasive procedure (reference standard). 32/504 samples failed during sequencing process (no gNIPT result), including 14 samples failed sequencing quality criteria and 18 samples failed libraries. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To validate the diagnostic accuracy of a gNIPT for detecting T21, T18 and T13 for a population in Germany and Switzerland. | ||
| Funding source or sponsor of the study | Study funded by LifeCodexx AG and GATC Biotech AG. | ||
| Informations about the authors contacted | Author was contacted on: 22 February 2016, 24 February and 19 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | No | ||
| High | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 200 pregnant women. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer. Fetal fraction DNA: not reported. 1) Positive for T21 and T18 if T score > 5. 2) Positive for T13 if T score > 4. 3) Positive for 45,X if T score for chrom. X > 0.04 and for chrom. Y < 0.04. In‐house test. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To estimate the feasibility of using a next‐generation sequencing technique for the noninvasive prenatal diagnosis of fetal aneuploidies. | ||
| Funding source or sponsor of the study | Funding source not reported. | ||
| Informations about the authors contacted | Author was contacted on: 9 September and 4 October 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: retrospective study from a prospective cohort. | ||
| Patient characteristics and setting | Number enrolled: 918 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. Biochemical serum‐screening results were reported in the study but 2 x 2 tables could not be derived. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. T13 were also assessed but the only case found was without follow‐up. SCA were also screened but inappropriate reference standard for the present review was used. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). 9/910 samples without follow‐up were excluded (all samples had positive gNIPT result). 2/9 women had abortion and 7/9 women declined invasive testing. 21/910 samples failed to give a risk score for gNIPT of the first blood samples including 1 haemolysed sample, 8 samples thawing due to transport delay, 3 due to cell‐free DNA extraction failures and 9 samples had low fetal fraction. 16/21 samples were repeated with new sampling. 14/16 samples obtained a gNIPT results and 2/16 samples failed to provide informative results and were classified as test failures. 7/910 samples failed during sequencing process (no gNIPT result). 5/7 samples failed the initial MPSS testing and were not resequenced and 2/7 samples failed the second MPSS testing. | ||
| Comparative | |||
| Aim to study | To report the initial clinical performance of gNIPT in detecting fetal chromosomal aneuploidies, especially T21, T18 and T13, in singleton pregnancies in Korea. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI performed sequencing and analysis. Study funded by Seoul Clinical Laboratories Research Grant (2015, President: Kyoung‐Ryul Lee). | ||
| Informations about the authors contacted | Author was contacted on: 13, 19 and 26 September 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| High | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, retrospective clinical evaluation study. | ||
| Patient characteristics and setting | Number enrolled: 1100 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 or HiSeq 2500 sequencer in multiplex. Mean fetal fraction DNA (± SD): low‐risk group: 10% (± 3.1%), high‐risk group (< 35 years): 11.9% (± 4.8%), and high‐risk group (≥ 35 years): 11.1% (± 3.4%). Median fetal fraction DNA (range): low‐risk group: 10.7% (3.1% to 22.9%), high‐risk group (< 35 years): 10.7% (4.9% to 28.3%), and high‐risk group (≥ 35 years): 11% (3.1% to 25.5%). | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To demonstrate the clinical performance of a simplified, low coverage, low cost MPSS assay (VisibiliT™) that combines a maternal age‐based risk for T21, T18, and T13, the fractional concentration of fetal DNA, and the representation of chromosomes 21, 18, and 13 in the sample to provide a risk score for T21, T18 and T13, with classification of fetal sex result. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 595 pregnant women. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex with FORTE algorithm. Mean fetal fraction DNA (± SD; range): 11.1% (± 4.1%; 4% to 30%). | ||
| Target condition and reference standard(s) | Target condition: T21. | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior the invasive procedure (reference standard). 51/520 samples failed the initial TMPS testing. 51/51 samples were repeated with a second aliquot of the first sampling and 35/51 samples obtained a gNIPT results. 16/520 samples failed during sequencing process (no gNIPT result), including 7 samples with low fetal DNA fraction and 9 samples failed laboratory processing or specimen issues. | ||
| Comparative | |||
| Aim to study | To evaluate the performance of a directed gNIPT method of ccfDNA analysis for fetal T21 by shipping the whole blood samples from Europe to a laboratory in the USA. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. 2 authors are paid employees of Ariosa Daignostics. 1 author is a board member of Ariosa Diagnostics. | ||
| Informations about the authors contacted | Author was contacted on: 22 April 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 136 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer with NIFTY™ algorithm. Fetal fraction DNA: not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. T13 was also assessed but no case was found. 45,X was also screened but inappropriate reference standard for the present review was used for pregnant women with gNIPT negative result. gNIPT data from 45,X were not shown in this review. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate the value of maternal plasma ccfDNA examination in detection of fetal chromosomal aneuploidy in pregnant women at advanced maternal age during the first trimester of pregnancy. | ||
| Funding source or sponsor of the study | Study not funded by industry. Study funded by National Science & Technology Pillar Program during the Twelfth Five‐year Plan Period (2012BA131B06). | ||
| Informations about the authors contacted | Author was contacted on: 19 May and 27 June 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 917 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 flow cell on a HiSeq sequencer. Fetal fraction DNA: not reported. 1) for T21, T18 and T13, positive if Z score > 3. 2) for 47,XXY and 47,XYY, positive if Z score Chrom. X > ‐3 and Z score Chrom. Y < 3. 3) for 45,X and 47,XXX, positive if Z score Chrom. X between ‐3 and 3 without Chrom. Y representation. | ||
| Target condition and reference standard(s) | Target conditions: T21 and T18. T13 was also assessed but no case was found. SCA was also assessed but inappropriate reference standard for the present review was used. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To investigate the clinical efficiency of gNIPT identifying fetal chromosomal aneuploidies. | ||
| Funding source or sponsor of the study | Study not funded by industry but Berry Genomics Co. Ltd give technical support. Study funded by the Community Development Fund, granted by the Department of Family Planning and Healthcare, Jiangsu Province, China. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
| Study characteristics | |||
| Patient sampling | Study design: retrospective study. | ||
| Patient characteristics and setting | Number enrolled: 5950 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex with NIFTY™ algorithm. Fetal fraction DNA: amount measured but not reported. 1) positive if t score ≥ 2.5 for autosomes. 2) positive if t score for Chrom. X < ‐2.5 for female fetuses for 45,X. 3) positive if t score for Chrom. X > 2.5 for female fetuses for 47,XXX. 4) positive if t score for Chrom. X > 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X (expected value of zero) for 47,XXY. 5) positive if t score for Chrom. X > 2.5 and R‐value (the ratio of the fetal DNA fraction estimated by chromosome Y to that estimated by chromosome X) between 1.8 and 2.2 for 47,XYY. Commercial test: BGI‐Shenzhen's prenatal test. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13. 45,X, 47,XXY, 47,XYY and 47,XXX were also screened but inappropriate reference standard for the present review was used. gNIPT data from SCA were not shown in this review. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard). No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the performance of a MPSS in detecting fetal sex chromosome aneuploidy (SCA) and to present a comprehensive clinical counselling protocol for SCA‐positive patients. Author also assessed autosomes aneuploidies. | ||
| Funding source or sponsor of the study | Funding source not reported but many authors are employees of the Clinical Laboratory of BGI Health, BGI‐Shenzen or of the Shenzen Birth Defect Screening Projet Lab. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 87 pregnant women. | ||
| Index tests | gNIPT by MPSS on Illummina Hiseq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. No other cutpoint reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18. 45,X and 47,XXX were also screened but inappropriate reference standard for the present review was used. | ||
| Flow and timing | It is not reported if the blood samples were collected before or after invasive procedure (reference standard). No repeated test reported. | ||
| Comparative | |||
| Aim to study | To evaluate the efficacy of using gNIPT technology in screening T21 among women of advanced maternal age and to provide evidence for prenatal screening of T21. | ||
| Funding source or sponsor of the study | Funding source not reported. | ||
| Informations about the authors contacted | Author was contacted on: 7 September 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 306 pregnant women. Number available for 2 x 2 tables: 301 pregnant women in the pilot validation set (subgroup of 98%). See Zhou 2014b for the integration set. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. No failed sample reported. No repeated test reported. | ||
| Comparative | |||
| Aim to study | To report the clinical application of gNIPT to detect chromosomal aneuploidies, especially T21, T18 and T13 in Chinese singleton pregnancies. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI‐Shenzhen made sequencing and analysis. Some authors are employees of BGI‐Shenzhen. | ||
| Informations about the authors contacted | Author was contacted on: 31 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. | ||
| Patient characteristics and setting | Number enrolled: 7705 pregnant women. Number available for 2 x 2 tables: 3950 pregnant women in the integration set (subgroup of 51%). | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. | ||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 141/7705 samples failed the initial MPSS testing. 141/141 samples were repeated with a new sampling and 137/141 samples obtained a gNIPT results. 4/7705 samples failed the second MPSS testing for low fetal fraction DNA (no gNIPT result). 3751/7701 samples without birth outcome were excluded (no reference standard). | ||
| Comparative | |||
| Aim to study | To report the clinical application of gNIPT to detect chromosomal aneuploidies, especially T21, T18 and T13 in Chinese singleton pregnancies. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI‐Shenzhen made sequencing and analysis. Some authors are employees of BGI‐Shenzhen. | ||
| Informations about the authors contacted | Author was contacted on: 31 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
CVS: chorionic villi sampling
DANSR: digital analysis of selected regions
FISH: fluorescence in situ hybridisation
gNIPT: genomics‐based non‐invasive prenatal testing
MAD: Median absolute deviation
MPSS: massively parallel shotgun sequencing
NCV: normalised chromosome value
SD: standard deviation
SNP: single nucleotide polymorphism
TMPS: targeted massively parallel sequencing
Characteristics of excluded studies [ordered by study ID]
Jump to:
| Study | Reason for exclusion |
| Not a diagnostic test accuracy study. Poster abstract. | |
| Decision making study. Observational study. Not a diagnostic test accuracy study. | |
| Samples overlap with Brady 2016. Most gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract of the 33rd Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Observational study with incomplete follow‐up. Samples overlap with Beamon 2014. | |
| Observational study with incomplete follow‐up. Not a diagnostic test accuracy study. Some gNIPT results unconfirmed by a reference standard test. | |
| Most women (95%) completed a questionnaire to report their birth outcome (inappropriate reference standard for this review). | |
| Tribune. Not a diagnostic test accuracy study. | |
| All samples overlap with Benachi 2015. | |
| Letter to the editor on Zhang 2015 without data. Not a diagnostic test accuracy study. | |
| Poster abstract of the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014. Patients with gNIPT negative result were without follow‐up (no reference standard). Incomplete 2 x 2 tables. | |
| Samples overlap with Bianchi 2012. Data excluded to avoid double counting. | |
| Editorial. Not a diagnostic test accuracy study. | |
| Data excluded to avoid double counting. Samples overlap with Bianchi 2015b. | |
| Not a diagnostic test accuracy study. Author presented some false positive cases in women with malignancy. | |
| Incomplete 2 x 2 table. In this observational study, most women (98.9%) had no follow‐up (no reference standard). | |
| Not a diagnostic test accuracy study. Author presented some false positive cases in women with malignancy. Samples overlap with Bianchi 2015a. | |
| Poster abstract. Not a diagnostic test accuracy study. Author presented some false positive cases in women with malignancy. Samples overlap with Bianchi 2015a. | |
| Technology Evaluation Center Assessment. Review. | |
| Review with new data but most gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract of the ISPD 17th International Conference on Prenatal Diagnosis and Therapy. Samples overlap with Huang 2014. | |
| Poster abstract. Samples overlap with Yeang 2014. | |
| Poster abstract. Samples overlap with Meck 2015. | |
| Incomplete 2 x 2 table. This letter presented women who had positive results after screening and were referred for invasive procedure to confirm the presence of fetal aneuploidy. Only, the true positive and false positive gNIPT results were reported. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Poster abstract. Samples overlap with Rabinowitz 2012a. | |
| Full poster from the 17th International Conference on Prenatal Diagnosis and Therapy, ISPD 2013 received. Samples overlap with Cirigliano 2014 and Ordoñez 2015. Insufficient information to derive 2 x 2 tables. | |
| Full poster from the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014 received. Samples overlap with Cirigliano 2013 and Ordoñez 2015. Insufficient information to derive 2 x 2 tables. | |
| Review with simulation model for gNIPT. | |
| Poster abstract of the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014. Samples overlap with Dar 2014. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results completed a questionnaire to report their birth outcome (inappropriate reference standard for this review). | |
| Implementation study. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). Some women had follow‐up by telephone (inappropriate reference standard for this review). | |
| Case report (deletion). | |
| Poster abstract. Retrospective observational study. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract. Some gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract. Insufficient information to derive 2 x 2 tables (gNIPT positive results only). Decision making. Samples overlap with Dobson 2016. | |
| Insufficient information to derive 2 x 2 tables (gNIPT positive results only). Decision making. | |
| Sequencing not based on maternal plasma ccfDNA. | |
| Method development. Analytical accuracy. Not a diagnostic test accuracy study. | |
| Editorial comment without new data. | |
| Review. Not a diagnostic test accuracy study. | |
| Poster abstract. All gNIPT results (TMPS) were confirmed with a previous gNIPT result (MPSS) (inappropriate reference standard for this review). Insufficient information to derive 2 x 2 tables. | |
| Poster abstract of the 8th European Cytogenetics Conference. Samples overlap with Faas 2012. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Observational study. Incomplete 2 x 2 data. Most patients were without follow‐up (no reference standard). | |
| Conference abstract of the 17th International Conference on Prenatal Diagnosis and Therapy, ISPD 2013. Samples overlap with Fairbrother 2013a. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Insufficient information to derive 2 x 2 tables. | |
| Not a diagnostic test accuracy study (implementation study). | |
| Poster abstract. All samples overlap with Fiorentino 2016. | |
| Poster abstract of the 35th Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Observational study. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). | |
| Observational study with incomplete follow‐up. Incomplete 2 x 2 table. Many gNIPT results unconfirmed by a reference standard test. | |
| Conference abstract. Proof‐of‐concept. | |
| Full poster from the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). | |
| Poster abstract. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract of the 33rd Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Samples overlap with Xiong 2015. | |
| Poster abstract of the 34th Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Samples overlap with Xiong 2015. | |
| Women were asked to complete a questionnaire to report their birth outcome (inappropriate reference standard for this review). Insufficient information to derive 2 x 2 tables. | |
| Most patients with gNITP negative result were without follow‐up (no reference standard) because 962 women had not yet delivered at the time of writing the publication. Insufficient information to derive 2 x 2 tables. Some patients overlap with del Mar Gil 2014. | |
| Decision making including gNIPT accuracy data. All samples overlap with Gil 2016. | |
| Poster abstract. Samples overlap with Kurtser 2015. | |
| Not a diagnostic test accuracy study. No sequencing data. | |
| Observational study. Not a diagnostic test accuracy study. Full poster received from the authors. Poster of the 17th International Conference on Prenatal Diagnosis and Therapy, ISPD 2013. | |
| Data excluded to avoid double counting. Blinded DNA sequencing libraries were provided by Sequenom from their clinical trial cohort (NCT00877292) and were resequenced by LifeCodexx. | |
| Research letter. Samples overlap with Pescia 2017. | |
| In reply to Bianchi 2015a. Not a diagnostic test accuracy study. | |
| Poster abstract. Insufficient information to derive 2 x 2 tables. | |
| Implementation study. Not a diagnostic test accuracy study. | |
| Poster abstract. Samples overlap with Stumm 2014. | |
| Conference abstract. Insufficient information to derive 2 x 2 tables. | |
| Method development. Data were reanalysed by a new algorithmic approach of PraenaTest®. Not a diagnostic test accuracy study. | |
| Not a next generation sequencing publication. NIPT was ultrasound and serum biomarkers. | |
| Incomplete 2 x 2 tables. Only gNIPT positive results presented. | |
| Poster abstract. All samples overlap with Hui 2015b. | |
| Implementation study. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). | |
| Poster abstract. All samples overlap with Jackson 2014. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. Samples overlap with Palomaki 2012. | |
| Proof‐of‐concept study with unblinded samples. Not a diagnostic test accuracy study. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results were followed‐up by telephone (inappropriate reference standard for this review). | |
| Not a next generation sequencing method. | |
| Method development. Incomplete 2 x 2 table. Most patients were without follow‐up (no reference standard). | |
| Not a diagnostic test accuracy study. Simulation model. | |
| Not next generation sequencing method. | |
| Methodological publication. Not a diagnostic test accuracy study. | |
| Poster abstract. Some gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. | |
| Methodological publication. Not a diagnostic test accuracy study. | |
| Bioinformatic simulation with Palomaki 2011 data. | |
| Method development (proof‐of‐concept study). Development of an advanced fetal fraction estimation method and aneuploidy determination algorithm. Not a diagnostic test accuracy study. | |
| Most patients with gNIPT negative result were without follow‐up (no reference standard). Incomplete 2 x 2 tables. | |
| Samples overlap with Palomaki 2011 and Palomaki 2012. Data excluded to avoid double counting. | |
| Poster abstract. Implementation study. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone or by email (inappropriate reference standard for this review). All samples overlap with Lau 2014. | |
| Sample overlap with Lau 2014. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone or by email (inappropriate reference standard for this review). | |
| Incomplete 2 x 2 table. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Poster abstract. Observational study about discordant gNIPT results. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract. Incomplete 2 x 2 table. | |
| Poster abstract. Incomplete 2 x 2 table. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. Samples overlap with Zimmermann 2013. | |
| Methodological publication about relation between fetal fraction and multiple clinical factors. | |
| Observational study. Unavailable information about gNIPT approach used. It is unclear if patients with gNIPT negative result were followed up (no reference standard). | |
| Not a diagnostic test accuracy study. No aneuploid case. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| This is a poster abstract. The full publication was also excluded. See Liao 2014 for reasons of exclusion. | |
| Incomplete 2 x 2 table for the retrospective and the prospective cohort. In prospective cohort, most patients were without follow‐up (no reference standard). For the retrospective cohort, number of gNIPT results was not reported. Sensitivity and specificity were presented for the retrospective cohort but 2 x 2 tables could not be derived. | |
| Letter to the editor about Bianchi 2014b without new data. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone (inappropriate reference standard for this review). | |
| Bioinformatic development. Comparison of sensitivity and specificity using 3 different count normalisation methods. | |
| Poster abstract. Bioinformatic development. Comparison of sensitivity and specificity using 3 different count normalisation methods. Samples overlap with Lo 2014. | |
| Samples overlap with Palomaki 2012. Data excluded to avoid double counting. | |
| Full poster. Observational study. Not a diagnostic test accuracy study. | |
| Samples overlap with Ma 2016. | |
| Poster abstract. All samples overlap with Ma 2016. | |
| Insufficient information to derive 2 x 2 tables. Women without invasive testing results were encouraged to report birth outcomes through the insurance policy reimbursed (inappropriate reference standard for this review). | |
| Poster abstract. Implementation study. Not a diagnostic test accuracy study. Insufficient information to derive 2 x 2 tables. | |
| Observational study. Not a diagnostic test accuracy study. | |
| Poster abstract. Samples overlap with Mazloom 2013. | |
| Incomplete 2 x 2 table. The clinician of women who passed gNIPT was encouraged to send ad hoc feedback to the lab (inappropriate reference standard for this review). | |
| Poster abstract. Incomplete 2 x 2 table. Samples overlap with McCullough 2014. | |
| Full poster of the 19th International Conference on Prenatal Diagnosis and Therapy, ISPD 2015. Most gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 × 2 tables. Some patients overlap with McCullough 2014. | |
| Most patients with gNIPT negative result were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. | |
| Poster abstract. Samples overlap with Meck 2015. | |
| Not a diagnostic test accuracy study. Observational study. | |
| Poster abstract. Samples overlap with Meck 2015. | |
| Review without original data. | |
| gNIPT negative results unconfirmed by a reference standard test. Not a diagnostic test accuracy study. | |
| Not a diagnostic test accuracy study. This these explore traditional screening tests. | |
| Health Technology Assessment. Not a diagnostic test accuracy study. | |
| Health Technology Assessment. Not a diagnostic test accuracy study. | |
| Poster abstract. Samples overlap with Norton 2015. | |
| Poster abstract. Samples overlap with Hooks 2014 and Nicolaides 2014a. | |
| Not a gNIPT method (other method). | |
| Not with ccfDNA (other sampling). | |
| Not a gNIPT method (other method). | |
| Completed clinical trial but no published data. | |
| Incomplete 2 x 2 data (ongoing study with cases only). | |
| Not a gNIPT method (other method). | |
| Incomplete 2 x 2 data (ongoing study with cases only). | |
| Observational study on gNIPT without fetal karyotype. | |
| Completed clinical trial but no published data. | |
| Completed clinical trial but no published data. | |
| Completed clinical trial but no published data. | |
| Not a diagnostic test accuracy study. | |
| Adult with T21. Not with pregnant women (other population). | |
| Not a diagnostic test accuracy study. | |
| Not with ccfDNA (other sampling). | |
| Incomplete 2 x 2 data. | |
| Completed clinical trial but no published data. | |
| Not a diagnostic test accuracy study. Pregnant women with gNIPT have not a reference standard. | |
| Inappropriate reference standard for this review (pregnancy outcome data obtained from the patient). | |
| Not a gNIPT method (other method). | |
| Observational study. Not a diagnostic test accuracy study. Incomplete 2 x 2 tables. Most gNIPT results unconfirmed by a reference standard test. | |
| Method validation for the NextSeq 500 platform. Not a diagnostic test accuracy study. | |
| Not a diagnostic test accuracy study. | |
| Poster abstract. All samples overlap with Nicolaides 2012. | |
| Simulation model on gNIPT implantation in first‐ or second‐tier test. | |
| Note on Nicolaides 2014a without new data. | |
| Target condition presented in this publication is not the focus of this review. Publication of next generation sequencing with ccfDNA for fetal triploidy. | |
| Full poster received from authors. Most patients were without follow‐up (no reference standard). Incomplete 2 x 2 tables. | |
| Bioinformatic simulation. | |
| Poster abstract. Samples overlap with Norton 2015. | |
| Bioinformatic simulation. | |
| Editorial on Norton 2015 without new data. | |
| Author reply to comments from Sentilhes 2015 and Smith‐Bindman 2015 about Norton 2015 without new data. | |
| Simulation model to compare sequential and ccfDNA screening with data published in the literature. Not a diagnostic test accuracy study. | |
| Bioinformatic simulation. | |
| Most patients were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. | |
| Poster abstract. Insufficient information to derive 2 x 2 tables. | |
| Full poster received. Some gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. | |
| Samples overlap with Palomaki 2012 (samples in Palomaki 2011 have been reanalysed in Palomaki 2012). Study excluded to avoid double counting. | |
| Samples overlap with Palomaki 2012. Conference abstract about Palomaki 2012 data. | |
| Editorial on Palomaki 2011 without new data. | |
| Not a diagnostic test accuracy study. | |
| Note about Palomaki 2015. Not a diagnostic test accuracy study. | |
| Incomplete 2 x 2 tables. Some women with gNIPT negative results were followed up by telephone (inappropriate reference standard for this review). | |
| Follow‐up for gNIPT negative results was ensured by an inquiry of two sets of randomly selected samples (inappropriate reference standard for this review). | |
| Not a next generation sequencing publication. NIPT was ultrasound measurement and serum biomarkers. | |
| Most patients with gNIPT negative result were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. | |
| Reply to Grati 2014 without sequencing data. | |
| Poster abstract. Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Poster abstract with incomplete 2 x 2 tables. | |
| Poster abstract. Samples overlap with Rabinowitz 2012a. | |
| Poster abstract. Samples overlap with Pergament 2014. | |
| Poster abstract. Sample overlap with Pergament 2014. | |
| Implementation study without sequencing data presented. | |
| Incomplete 2 x 2 tables. Most patients were without follow‐up (no reference standard). | |
| Poster abstract. Samples overlap with Bianchi 2012. | |
| Methodological publication about fetal DNA fraction with MELISSA samples. | |
| Insufficient information to derive 2 x 2 tables. | |
| Insufficient information to derive 2 x 2 tables. | |
| Conference abstract from the 19th International Conference on Prenatal Diagnosis and Theraphy, ISPD 2015. Simulation model. Not a diagnostic test accuracy study. | |
| Not a diagnostic test accuracy study. All samples overlap with Gil 2016 and Quezada 2015. | |
| Method development of version 2 to SNP‐based gNIPT. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept of fetal fraction quantification by qPCR. | |
| Most patients were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. Samples overlap with Sago 2015. | |
| Some women reported their birth outcome (inappropriate reference standard for this review). Information about false positive results were insufficient to derive all 2 x 2 tables. | |
| Poster abstract. Bioinformatics development with unblinded samples. | |
| Poster abstract. Incomplete 2 x 2 table. | |
| Poster abstract. Samples overlap with Bianchi 2014b. | |
| Comment about Norton 2015 without new data. | |
| Women with gNIPT result were without follow‐up at birth. | |
| Full poster received. Insufficient information to derive 2 x 2 tables. Some gNIPT results unconfirmed by a reference standard test. | |
| Simulation model. Not a diagnostic test accuracy study. | |
| Poster abstract. Full poster not received. Incomplete 2 x 2 table. | |
| Poster abstract about patient perceptions of gNIPT from the multi‐centered Canadian PEGASUS trial. gNIPT results compared with first trimester combined test (inappropriate reference standard for this review). | |
| Poster abstract. Samples overlap with Shaw 2014. | |
| Method development. Not a diagnostic test accuracy study. | |
| Incomplete 2 x 2 table. gNIPT negative result unconfirmed by a reference standard test. | |
| Poster abstract. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without a reference standard test. | |
| Letter to the editor on Bianchi 2015a without data. | |
| Comment about Norton 2015 without new data. | |
| Poster abstract. Some samples overlap with Sparks 2012a. | |
| Method development (all unblinded samples). Incomplete 2 x 2 table. Most patients with gNIPT negative result were unconfirmed by a reference standard test. | |
| Poster abstract. Samples from MELISSA study (potentially overlap). | |
| Not a next generation sequencing method. | |
| Women were followed up by telephone interview to find out their birth outcome (inappropriate reference standard for this review). | |
| Proof‐of‐concept. | |
| Incomplete 2 x 2 table. Only women with gNIPT positive result were reported. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Poster abstract. Samples overlap with Stumm 2014. | |
| Poster abstract. Samples overlap with Stumm 2014. | |
| Not a diagnostic test accuracy study. | |
| Publication about Bianchi 2012 without new data. | |
| Not a diagnostic test accuracy study. Simulation model. | |
| Women with gNIPT negative results were followed up by telephone interview (inappropriate reference standard for this review). Insufficient information to derive 2 x 2 tables. | |
| Incomplete follow‐up. Incomplete 2 x 2 table. Many gNIPT results unconfirmed by a reference standard test. | |
| Most patients with gNIPT negative result were without reference standard test. Providers were encouraged to report discordant clinical outcomes. Insufficient information to derive 2 x 2 tables. | |
| gNIPT results unconfirmed by a reference standard test. Their reference method is verifi® results (inappropriate reference standard for this review). | |
| Not a diagnostic test accuracy study. Observational study and decision making about gNIPT uptake in their center. | |
| Full poster received. Internal verification set and implantation in their centre. Not a diagnostic test accuracy study. | |
| Not a next‐generation sequencing method with ccfDNA. | |
| Poster abstract. Insufficient information to derive 2 x 2 tables. | |
| Poster abstract. Samples overlap with Valderramos 2016c. | |
| Insufficient information to derive 2 x 2 tables. Retrospective cohort of patients with gNIPT positive results. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Simulation model. Not a diagnostic test accuracy study. | |
| Poster abstract. All samples overlap with Verweij 2013. | |
| Not a diagnostic test accuracy study. Prenatal screening workflow proposed. | |
| Not a diagnostic test accuracy study. Prenatal screening workflow proposed. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone (inappropriate reference standard for this review). | |
| Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Editorial on Wang 2015b without new data. | |
| Not a diagnostic test accuracy study. | |
| Full poster received. Observational study and incomplete follow‐up. | |
| Simulation model for gNIPT implantation. Not a diagnostic test accuracy study. | |
| Commentary about gNIPT for microdeletion syndromes and rare autosomal trisomies. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Incomplete 2 x 2 table. Women with gNIPT negative results were followed‐up by telephone (inappropriate reference standard for this review). | |
| Poster abstract. Incomplete 2 x 2 table. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Proof‐of‐concept. Not a diagnostic test accuracy study. | |
| Implementation assessment of 2 sequencing platforms for gNIPT in a routine clinical environment. Not a diagnostic test accuracy study. |
ccfDNA: circulating cell‐free DNA
gNIPT: genomics‐based non‐invasive prenatal testing
MPSS: massively parallel shotgun sequencing
TMPS: targeted massively parallel sequencing
Characteristics of ongoing studies [author‐defined order]
Jump to:
| Trial name or title | Publication's title: False positive and false negative results of cell free DNA testing. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. |
| Index and comparator tests | gNIPT by TMPS or MPSS by commercial company providing gNIPT in Turkey (Ariosa Diagnostics, Inc., BGI‐Shenzhen, Illumina, Inc, Natera, Inc. and Sequenom, Inc). |
| Starting date | Not reported. |
| Contact information | Dr Seher Basaran Department of Medical Genetics Istanbul University, Istanbul Medical Faculty TURKEY 90 (212) 4142000 |
| Aim to study | To demonstrate the importance of confirmation of fetus genotype by invasive testing after gNIPT. |
| Funding source or sponsor of the study | The genetic centre is not affiliated with any commercial company providing gNIPT. |
| Information about the authors contacted | Author was contacted on: 12, 14 and 18 January 2016. |
| Notes | At the time of this writing, the authors plan to publish a full publication soon. |
| Trial name or title | Poster's title: Actual rates of recommended diagnostic testing after first‐trimester screening vs same‐day screening by cell free DNA. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. |
| Index and comparator tests | MPS. |
| Starting date | January to June 2015. |
| Contact information | Susan Klugman Department of Obstetrics and Gynecology and Women's Health, Albert Einstein College of Medicine, Montefiore Medical Center 1695 Eastchester Road, Bronx, NY 10461, United States. |
| Aim to study | To compare actual patient referrals for post‐screen diagnostic tests following first‐trimester screening vs same day ccfDNA. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 1 and 23 September 2016. |
| Notes | Authors are working on data at the time of writing and they plan to submit for publication. |
| Trial name or title | Oral presentation's title: Noninvasive prenatal diagnosis of fetal aneuploidy by massively parallel sequencing of maternal plasma DNA. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and SCA. |
| Index and comparator tests | gNIPT by MPSS on Illumina GAIIx/HiSeq 2000 sequencer. |
| Starting date | Not reported. |
| Contact information | Fang Chen, Beijing Genomics Institute, Shenzhen, China |
| Aim to study | To assess gNIPT with ccfDNA performance on fetal aneuploidies. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | BGI was contacted on: 19 May 2016. |
| Notes | Cohort of 5268 pregnant women. They successfully identified 62 cases of T21, 40 cases of T18, 3 cases of T13, 13 cases of SCA. In a cohort of karyotyping cases, the sensitivity and specificity of the aneuploidy fetus detection was 100% and 100%, respectively. |
| Trial name or title | Abstract's title: Non‐Invasive prenatal testing for the most common aneuploidies (trisomies 21, 18, and 13) using a semiconductor‐sequencing platform: a French multicenter pilot study. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. |
| Index and comparator tests | gNIPT on semiconductor sequencing platform (MPSS). |
| Starting date | Not reported. |
| Contact information | J.P. Da Fonseca, Inserm U1016 Plateforme Génomique, Paris, France. |
| Aim to study | To validate a common protocol and to evaluate the efficiency and reliability of gNIPT of the most common chromosomal aneuploidies using a semiconductor sequencing platform. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 19 January 2016 and 23 March 2016. |
| Notes | Conference Abstract of the 10th European Cytogenetics Conference of the European Cytogenetics Association, ECA 2015. Prospective study of 500 pregnant women at high risk of fetal aneuploidy who undergo fetal karyotyping. The NIPT results matched the fetal karyotyping results in all of the cases: all trisomies were detected. |
| Trial name or title | Comparison of false positive rates in prenatal combined screening and cell free DNA screening for trisomy 21 (ReFaPo study). |
| Target condition and reference standard(s) | Target condition: T21. Reference standard: prenatal or postnatal karyotype. |
| Index and comparator tests | gNIPT. |
| Starting date | July 2016. |
| Contact information | Karl Oliver Kagan University of Tuebingen |
| Aim to study | To compare the false positive rate of cell‐free DNA and traditional screening methods in a randomised controlled trial in a cohort without prior risk of fetal aneuploidy. |
| Funding source or sponsor of the study | Study funded by CENATA GmbH who does the analysis. |
| Information about the authors contacted | No need for further contact. |
| Notes | Target number of participants: 1400. Recruitment end date: March 2017. Intention to publish date: October 2018. DOI 10.1186/ISRCTN11174071 |
| Trial name or title | Clinical implementation of noninvasive prenatal testing in twin pregnancies with assisted reproductive technique treatment. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype or clinical outcomes. |
| Index and comparator tests | gNIPT by MPSS. Commercial test: BGI Shenzhen’s prenatal test. |
| Starting date | Not reported. |
| Contact information | BGI‐Shenzhen Shenzhen, China |
| Aim to study | To assess the clinical implementation of MPS‐based NIPT in twin pregnancies with assisted reproductive technique treatment. |
| Funding source or sponsor of the study | Not reported but BGI‐Shenzhen made sequencing and analyses. |
| Information about the authors contacted | No need for further contact. |
| Notes | Some women were still pregnant at the time of writing this poster abstract. |
| Trial name or title | Maternal non‐invasive fetal DNA test used in prenatal diagnosis. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standard: fetal karyotype of amniotic fluid. |
| Index and comparator tests | gNIPT by MPSS. NIFTY™ prenatal test by BGI‐Shenzhen. |
| Starting date | In 2012. |
| Contact information | Mu Y. Beijing United Family Hospital Beijing, China. |
| Aim to study | Not reported. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 19 April and 19 May 2016. No reply received from the author. |
| Notes | Poster abstract. Some women were still pregnant at the time of writing this poster abstract. |
| Trial name or title | Specimen collection from pregnant women at increased risk for fetal aneuploidy. |
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT. |
| Starting date | May 2011. |
| Contact information | Sequenom, Inc. |
| Aim to study | To develop a prenatal aneuploidy test using ccfDNA from blood samples from pregnant women who have an increased risk indicator/s for fetal chromosomal aneuploidy detection (T21). |
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | A safer pre‐natal diagnosis using free DNA in maternal blood (IONA®). |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and other chromosomal abnormalities yet to be determined. Reference standards: prenatal karyotype and follow‐up for 1 year. |
| Index and comparator tests | gNIPT by TMPS (selective amplification of fetal DNA) by Premaitha Health. |
| Starting date | April 2007. |
| Contact information | Brenda Kelly National Health Service, United Kingdom |
| Aim to study | To validate a novel gNIPT method that could increase the titre of fetal DNA within a given sample. |
| Funding source or sponsor of the study | Study funded by Premaitha Health. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | Prenatal Non‐invasive Aneuploidy Test Utilizing SNPs trial (PreNATUS). |
| Target condition and reference standard(s) | Target conditions: aneuploidy in a fetus at chromosomes 13, 18, 21, X and Y. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by TMPS (SNP based technology by Natera, Inc.). |
| Starting date | January 2012. |
| Contact information | Ronald Wapner, MD, Columbia University |
| Aim to study | To assess the diagnostic capability of an informatics enhanced SNP based technology (Parental Support) to identify pregnant women who are carrying a fetus with an aneuploidy from free floating DNA in the maternal blood. |
| Funding source or sponsor of the study | Study funded by Natera, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | Study of the efficacy of new non‐invasive prenatal tests for screening for fetal trisomies using maternal blood (PEGASUS). |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: prenatal or neonatal karyotype or medical record from birth. |
| Index and comparator tests | gNIPT by Semiconductor MPSS (Ion Torrent Proton™) or optical‐based MPSS (Illumina) or by TMPS with Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
| Starting date | August 2013. |
| Contact information | François Rousseau CHU de Québec Québec, Canada |
| Aim to study | To perform a pan‐Canadian large‐scale validation study comparing the relative effectiveness and clinical performances of 2 index gNIPT methods using fetal ccfDNA in maternal blood in Canadian clinical laboratories between themselves and with that of fetal karyotype for detecting fetal aneuploidy of chromosomes 13, 18 and 21 and to compare the accuracy of this new gNIPT method with traditional prenatal screening methods. |
| Funding source or sponsor of the study | Study funded by Centre Hospitalier Universitaire de Québec, Laval University, Genome Canada, Genome Quebec, Genome British Columbia and Canadian Institutes of Health Research (CIHR). |
| Information about the authors contacted | No need for further contact. |
| Notes | Recruitment of patients completed (near 5000 pregnant women enrolled). at the time of writing, they are sequencing 3600 pregnant women with the 2 gNIPT MPSS platforms. A subsample of about 2300 blood samples was analysed by Ariosa Diagnostics, Inc (TMPS). Estimated study completion date: June 2017. |
| Trial name or title | Non‐invasive Chromosomal Evaluation of Trisomy study (NICHE). |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by TMPS by Ariosa Diagnostics, Inc. |
| Starting date | April 2014. |
| Contact information | Romielle Aquino 408‐209‐9098 Or Thomas Musci 408‐229‐7500 |
| Aim to study | To provide clinically annotated samples to support continued improvements in the Ariosa Diagnostics, Inc Test content, methodology, specimen processing and quality control. |
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | Multiple gestation study. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and SCA. Reference standards: fetal karyotype (amniocentesis or CVS) or genetic testing from cheek swab or saliva from live‐born children. |
| Index and comparator tests | gNIPT by TMPS by Natera, Inc. |
| Starting date | March 2013. |
| Contact information | Brian Kirshon Houston Perinatal Associates Or Zach Demko Natera, Inc. |
| Aim to study | To demonstrate the accuracy of our new NATUS diagnostic method to determine the genetic health of the developing fetuses in a multiple gestation pregnancy from a maternal blood sample. |
| Funding source or sponsor of the study | Study funded by Natera, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | High risk multiple gestation study. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and SCA. Reference standards: fetal karyotype (amniocentesis or CVS) or genetic testing from cheek swab or saliva from live‐born children. |
| Index and comparator tests | gNIPT by TMPS by Natera, Inc. |
| Starting date | August 2014. |
| Contact information | Joanne Stone Mt. Sinai Hospital, New York |
| Aim to study | To demonstrate the accuracy of our proprietary algorithm method to determine the genetic health of the developing fetuses in a multiple gestation pregnancy from a maternal blood sample. |
| Funding source or sponsor of the study | Study funded by Natera, Inc., Mount Sinai Hospital New York, Montefiore Medical Center, Long Island Jewish Medical Center and Tufts Medical Center. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | Non‐invasive screening for fetal aneuploidy. |
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by MPSS by Progenity, Inc. |
| Starting date | March 2015. |
| Contact information | Richard Porreco Obstetrix Medical Group of Colorado |
| Aim to study | To detect whole chromosome abnormalities on all chromosomes 13, 16, 18, 21, X and Y, in the fetus through analysis of ccfDNA and compound sample DNA in maternal blood. |
| Funding source or sponsor of the study | Study funded by Progenity, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | T21,18 and 13 screening by cell free fetal DNA in low risk patients (DEPOSA). |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by MPSS. |
| Starting date | June 2015. |
| Contact information | Alexandra Benachi Antoine Béclère Hospital |
| Aim to study | To evaluate the performance of gNIPT in a population of pregnant women with and without in vitro fertilisation (IVF) concomitantly to regular first‐trimester trisomy 21 (T21) screening using maternal age, nuchal fold measurement and serum screening. |
| Funding source or sponsor of the study | Study funded by Assistance Publique ‐ Hôpitaux de Paris. |
| Information about the authors contacted | No need for further contact. |
| Notes | Recruitment of patients completed (933 pregnant women enrolled). |
| Trial name or title | Expanded Noninvasive Genomic Medical Assessment: the Enigma study. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, microdeletion syndromes, sex chromosome abnormalities, infectious and other diseases, and blood group typing. Reference standard: fetal karyotype or medical records. |
| Index and comparator tests | gNIPT by MPSS provided by Progenity, Inc. |
| Starting date | October 2015. |
| Contact information | Paul Bien 760‐494‐1743 |
| Aim to study | To evaluate the relative clinical sensitivity, specificity, and performance of the laboratory‐developed test as a screening test for fetal chromosomal aneuploidy, infectious and other diseases, and RhD genotyping in the general population of pregnant women. |
| Funding source or sponsor of the study | Study funded by Progenity, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | Nationwide demonstration project of next‐generation sequencing of cell‐free DNA in maternal plasma in Japan: 1‐year experience |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. |
| Index and comparator tests | gNIPT by MPSS. |
| Starting date | 15 November 2012. Recruitment period between April 2013 to March 2014. |
| Contact information | Haruhiko Sago National center for Child‐health and development Perinatal Center 2‐10‐1Ookura, Setagaya‐ku, Tokyo 03‐3416‐0181 sagou‐[email protected] |
| Aim to study | To evaluate the quality of the genetic counselling in Japan. Sago 2015 reported the 1‐year experience of a nationwide demonstration project to introduce gNIPT of fetal aneuploidy from maternal plasma and discuss how to implement this program in Japan. |
| Funding source or sponsor of the study | Study supported by the Grant of the National Center for Child Health and Development 24‐3, Japan. Sequenom, Inc made gNIPT. |
| Information about the authors contacted | Author was contacted on: 6 April and 14 June 2016. |
| Notes | Authors continue collecting follow‐up data in the study population. |
| Trial name or title | Clinical implementation of non‐invasive prenatal study for detecting aneuploidies by fetal DNA based on single nucleotide polymorphisms: 2 years in Mexico. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. |
| Index and comparator tests | gNIPT by TPMS. Commercial test: Natera’s prenatal test. |
| Starting date | Recruitment period: March 2013 to February 2015. |
| Contact information | Dr. Rafael Sánchez Usabiaga |
| Aim to study | To describe our experience of 2 years integrating gNIPT by ccfDNA in its variant of single nucleotide polymorphism (SNPs) as a screening method for the detection of common aneuploidies, since 9 weeks of gestation. |
| Funding source or sponsor of the study | Not reported but Natera, Inc. made gNIPT sequencing and analyses. |
| Information about the authors contacted | No need for further contact. |
| Notes | There are 270 pregnant women included in this study. |
| Trial name or title | TRIDENT: or monitored NIPT implementation in the Netherlands. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. |
| Index and comparator tests | gNIPT by MPSS. |
| Starting date | 01 April 2014. |
| Contact information | Dr. Erik Sistermans. VU University Medical Center +31‐20‐020‐4448346 |
| Aim to study | To investigate and evaluate all relevant aspects of the introduction of NIPT in the Dutch prenatal screening program. |
| Funding source or sponsor of the study | The TRIDENT study was designed and proposed by the national multidisciplinary NIPT consortium. |
| Information about the authors contacted | Author have been contacted on: 9 December 2015 and 15 March 2016. |
| Notes | Conference abstract presented at the Annual conference of the European Society of Human Genetics at Glasgow, Scotland, UK. http://www.emgo.nl/research/quality‐of‐care/research‐projects/1451/trident‐study‐trial‐by‐dutch‐laboratories‐for‐evaluation‐of‐non‐invasive‐prenatal‐testing‐nipt/background/ The authors plan to publish a full publication soon. |
| Trial name or title | Genetic non invasive prenatal testing: A clinical and technical experience of 3000 cases with follow‐up. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. |
| Index and comparator tests | gNIPT by MPSS. Commercial test: TrisoNIM® prenatal test by NIMGenetics Genomics. |
| Starting date | Not reported. |
| Contact information | Juan C Cigudosa NIMGenetics Genomics Madrid, Spain. |
| Aim to study | To show a NIPT protocol, called TrisoNIM®, which has been partially performed in our laboratory, based in massive parallel sequencing. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author were contacted on: 29 February, 22 March, 15 and 27 June 2016. |
| Notes | Full poster received from the authors. |
| Trial name or title | Introduction of noninvasive prenatal testing for fetal trisomies: preliminary results and consequences on invasive samplings. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Microdeletion syndromes can also be detected. |
| Index and comparator tests | gNIPT by MPSS. |
| Starting date | December 2013. |
| Contact information | Dr Van Wymersch Didier, |
| Aim to study | To analyse a year of gNIPT implantation in our institute and to analyse gNIPT implication in chromosomal abnormalities screening politic. |
| Funding source or sponsor of the study | No reported. Samples analysed at BGI. |
| Information about the authors contacted | Author was contacted on: 12 September 2016. |
| Notes | This publication showed the first 683 samples. At the time of writing, authors have a much larger population of 2132 pregnant women. No false negative results have been observed to date for all the pregnancies that have already come to term. |
| Trial name or title | The first 3000 Non‐Invasive Prenatal Tests (NIPT) with the Harmony test in Belgium and the Netherlands. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. |
| Index and comparator tests | gNIPT by TMPS. Commercial test: Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
| Starting date | Recruitment period: March 2013 to December 2013. |
| Contact information | Patrick Willems patrick.willems@genetic‐diagnostic.net |
| Aim to study | To report the results of the first 3000 consecutive gNIPT tests performed in pregnant women from Belgium and the Netherlands. |
| Funding source or sponsor of the study | Not reported. Ariosa Diagnostics, Inc made sequencing and analysis. |
| Information about the authors contacted | No need for further contact. |
| Notes |
| Trial name or title | Maternal non‐invasive fetal DNA test used in prenatal diagnosis. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. |
| Index and comparator tests | gNIPT by MPSS by BGI‐Shenzhen. |
| Starting date | Patients recruited in 2012. |
| Contact information | Yu M or Fei S. Beijing United Family Hospital. |
| Aim to study | To determine gNIPT accuracy in Chinese population. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 15 May 2016. BGI was contacted on: 19 May 2016. No reply received from the author or BGI. |
| Notes | Conference abstract. Some women were still pregnant at the time of writing their conference abstract. |
| Trial name or title | Prenatal detection of fetal aneuploidy on the Ion Torrent Proton™ platform. |
| Target condition and reference standard(s) | Target condition: T21. |
| Index and comparator tests | gNIPT by MPSS on the Proton™ platform. |
| Starting date | Not reported. |
| Contact information | Sequenom, Inc. |
| Aim to study | To examine the performance of a gNIPT for fetal aneuploidy on the Ion Torrent Proton™ platform. |
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. |
| Information about the authors contacted | Author was contacted on: 19 April and 15 June 2016. |
| Notes | Full poster received from authors. This study includes 156 samples including 16 women carrying a T21 fetus. All patient samples were correctly identified according to their karyotype results. |
CVS: chorionic villi sampling
gNIPT: genomics‐based non‐invasive prenatal testing
MPSS: massively parallel shotgun sequencing
TMPS: targeted massively parallel sequencing
Data
Presented below are all the data for all of the tests entered into the review.
| Test | No. of studies | No. of participants |
| 1 MPSS T21 Show forest plot | 41 | 50133 |
| Test 1  MPSS T21. | ||
| 2 MPSS T18 Show forest plot | 38 | 49003 |
| Test 2  MPSS T18. | ||
| 3 MPSS T13 Show forest plot | 29 | 46090 |
| Test 3 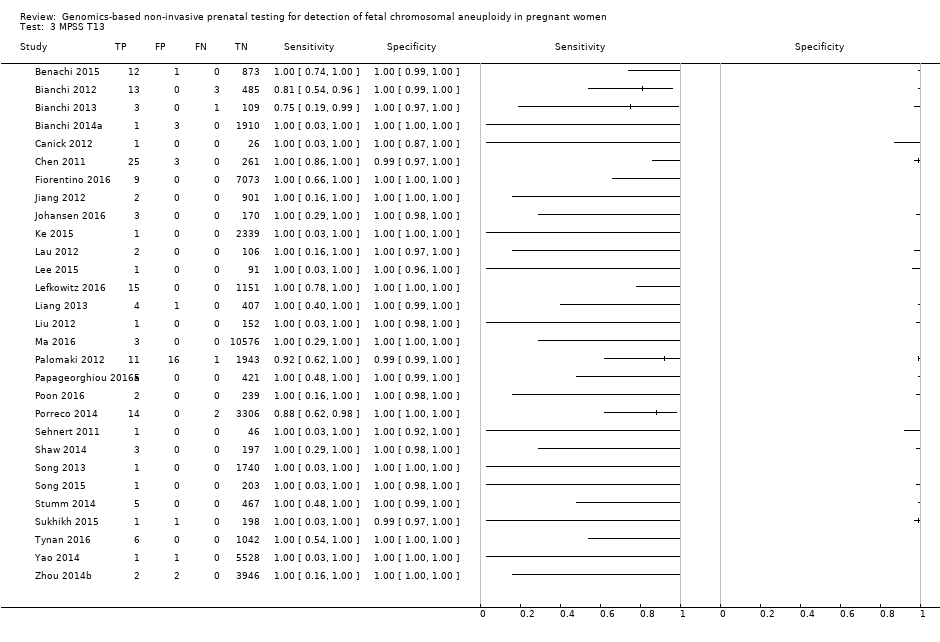 MPSS T13. | ||
| 4 MPSS 45,X Show forest plot | 14 | 7867 |
| Test 4  MPSS 45,X. | ||
| 5 MPSS 47, XXX Show forest plot | 5 | 5449 |
| Test 5  MPSS 47, XXX. | ||
| 6 MPSS 47,XXY Show forest plot | 8 | 6588 |
| Test 6  MPSS 47,XXY. | ||
| 7 MPSS 47,XYY Show forest plot | 8 | 6629 |
| Test 7 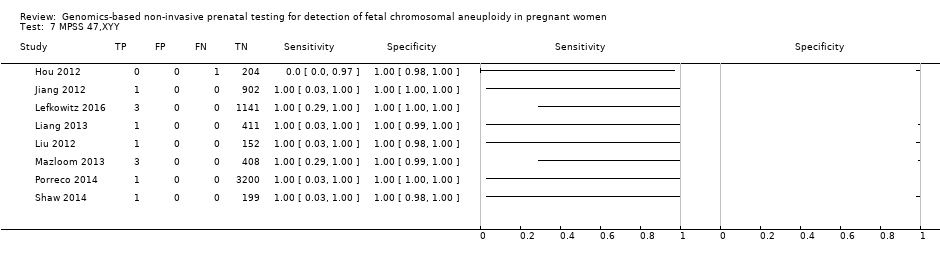 MPSS 47,XYY. | ||
| 8 MPSS all 7 aneuploidies Show forest plot | 44 | 50864 |
| Test 8  MPSS all 7 aneuploidies. | ||
| 9 MPSS, autosomes Show forest plot | 43 | 50453 |
| Test 9  MPSS, autosomes. | ||
| 10 MPSS, SCA Show forest plot | 14 | 7911 |
| Test 10 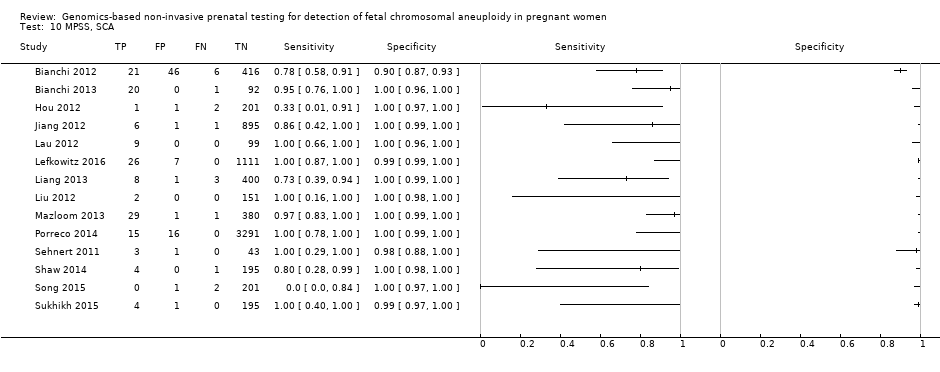 MPSS, SCA. | ||
| 11 TMPS T21 Show forest plot | 16 | 32487 |
| Test 11  TMPS T21. | ||
| 12 TMPS T18 Show forest plot | 12 | 30319 |
| Test 12  TMPS T18. | ||
| 13 TMPS T13 Show forest plot | 10 | 22868 |
| Test 13 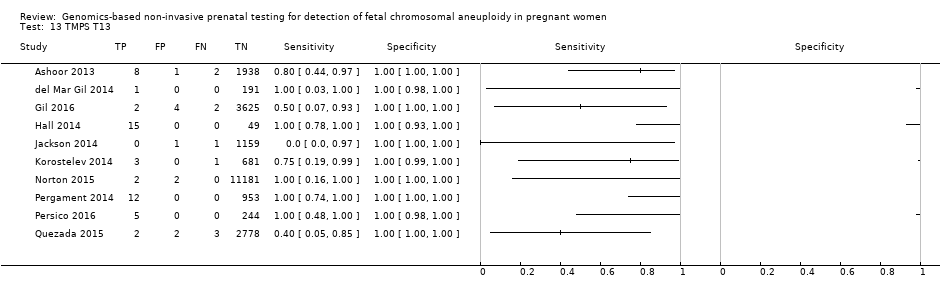 TMPS T13. | ||
| 14 TMPS 45,X Show forest plot | 6 | 2214 |
| Test 14 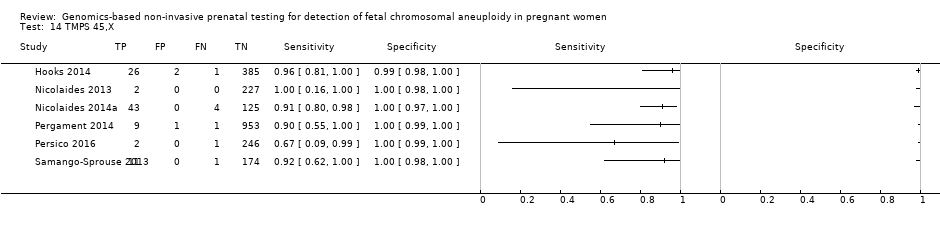 TMPS 45,X. | ||
| 15 TMPS 47,XXX Show forest plot | 2 | 586 |
| Test 15 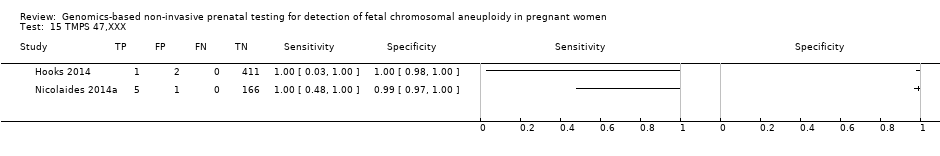 TMPS 47,XXX. | ||
| 16 TMPS 47,XXY Show forest plot | 4 | 1021 |
| Test 16  TMPS 47,XXY. | ||
| 17 TMPS 47,XYY Show forest plot | 2 | 358 |
| Test 17 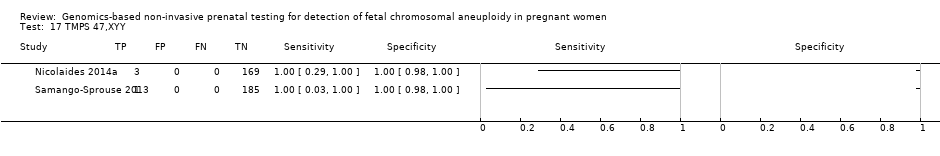 TMPS 47,XYY. | ||
| 18 TMPS all 7 aneuploidies Show forest plot | 21 | 35275 |
| Test 18  TMPS all 7 aneuploidies. | ||
| 19 TMPS, autosomes Show forest plot | 18 | 34473 |
| Test 19  TMPS, autosomes. | ||
| 20 TMPS, SCA Show forest plot | 6 | 2214 |
| Test 20  TMPS, SCA. | ||
| 21 Traditional screening tests, autosomes Show forest plot | 5 | 24279 |
| Test 21  Traditional screening tests, autosomes. | ||
| 22 Traditional screening tests T21 Show forest plot | 2 | 17753 |
| Test 22  Traditional screening tests T21. | ||
| 23 Traditional screening tests T18 Show forest plot | 2 | 17747 |
| Test 23 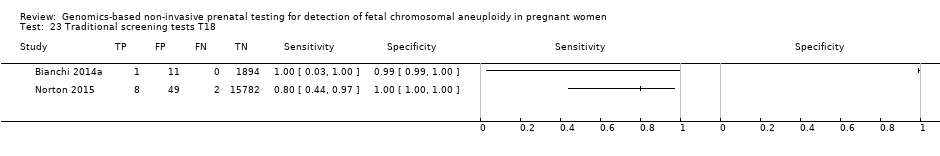 Traditional screening tests T18. | ||
| 24 Traditional screening tests T13 Show forest plot | 1 | 11185 |
| Test 24 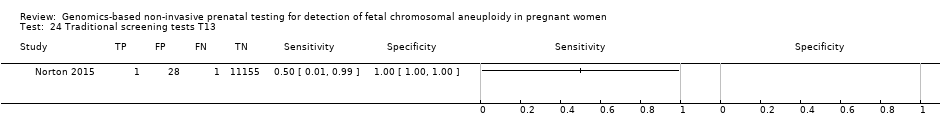 Traditional screening tests T13. | ||

Difference between massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS). Genomics‐based non‐invasive prenatal testing (gNIPT) aims to count the number of copies of DNA fragments from the chromosomes of interest (chromosome 21 (Chrom. 21) in this example) present in circulating cell‐free DNA (ccfDNA) from a pregnant woman, relative to a reference set of chromosomes (Ref. Chrom.). DNA fragments circulating in maternal blood in the case of a euploid (left) and aneuploid (right) pregnancy are illustrated (top). MPSS produces a large number of sequence reads from all chromosomes while TMPS generates a larger proportion of reads from the chromosomes of interest (bottom). In both methods, sequence reads can be used to detect a slight excess of fetal genomic material coming from the chromosome of interest. Figure was created by FR.
Current clinical pathway and three proposed uses of genomics‐based non‐invasive prenatal testing (gNIPT). Currently (on the left), pregnant women can have a prenatal screening test consisting of biomarkers or ultrasound, or both. For high‐risk pregnant women, an invasive diagnostic test (karyotyping) is offered. In the present review, we propose 3 different clinical pathways. First, gNIPT could be used as a triage test, to decide which pregnant women should receive further testing. Second, gNIPT could be used to replace current prenatal screening tests. Finally, gNIPT could be used to replace current invasive diagnostic tests (if diagnostic performance permits). At any point in a clinical pathway, a pregnant woman may decide not to proceed with other tests (not shown in the figure). Figure was designed by CL, JB, MB and YT.

PRISMA flow diagram for selection of studies from January 2007 to October 2016.
#: number, DTA: diagnostic test accuracy, NTIS: The National Technical Information Service and WHO ICTRP: World Health Organization International Clinical Trials Registry Platform.

Risk of bias and applicability concerns summary: review authors' judgements about each domain for each of the studies included for massively parallel shotgun sequencing (MPSS).
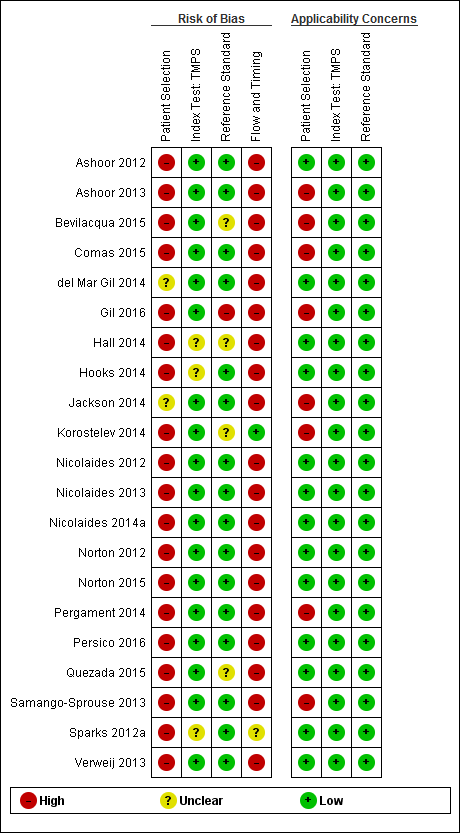
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each study included for targeted massively parallel sequencing (TMPS).
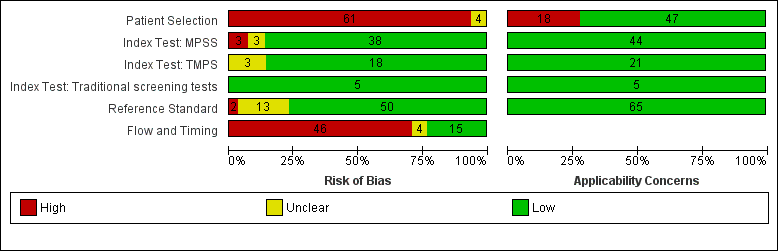
Risk of bias and applicability concerns (all tests included): review authors' judgements about each domains presented as percentages across included studies. MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing.
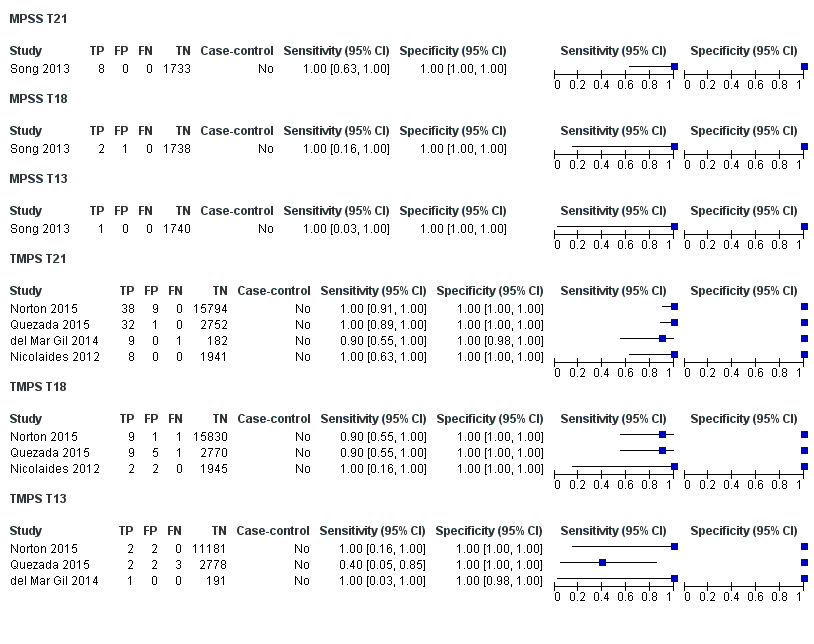
Forest plot of MPSS and TMPS for T21 in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
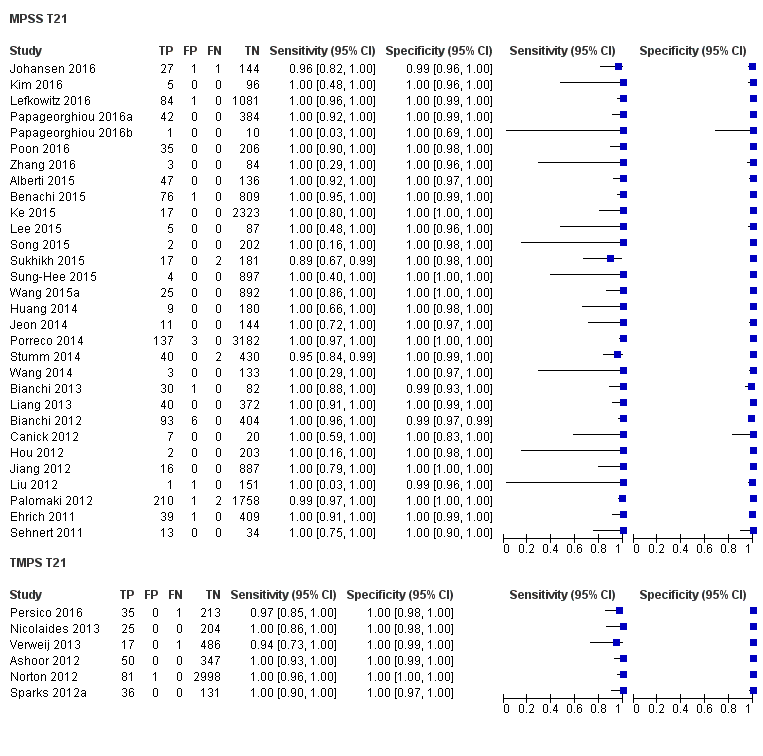
Forest plot of MPSS and TMPS for T21 in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
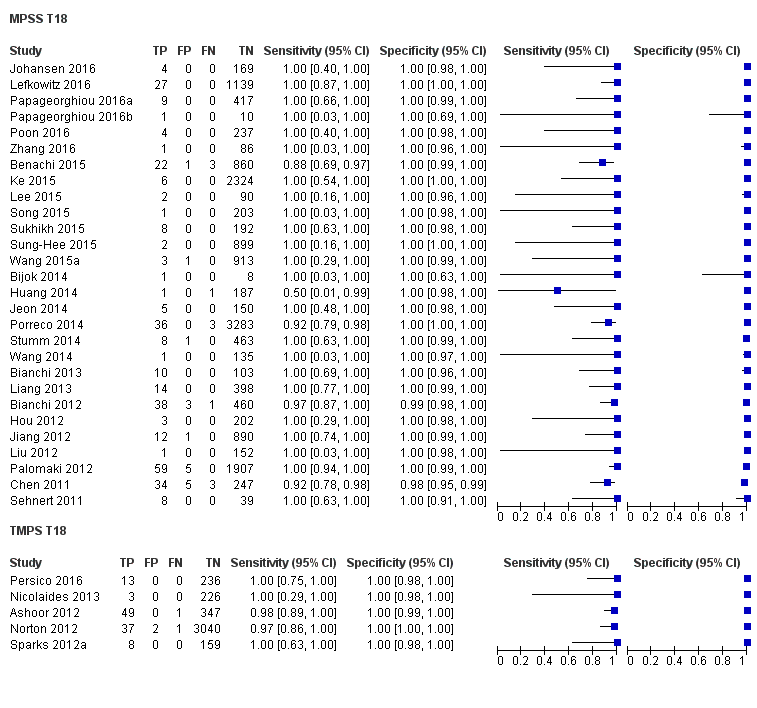
Forest plot of MPSS and TMPS for T18 in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for T13 in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for 45,X in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for 47,XXX, 47,XXY and 47,XYY in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for autosomes (T21, T18 and T13 combined) in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for autosomes (T21, T18 and T13) in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined) in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
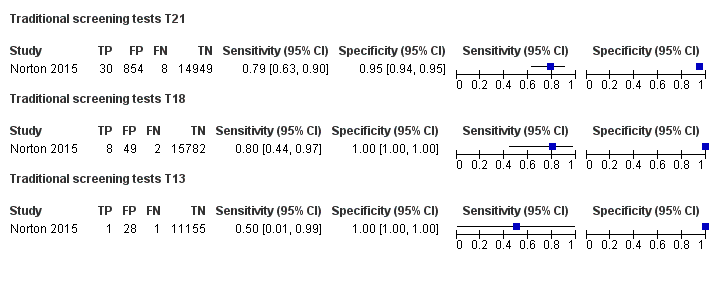
Forest plot of traditional screening tests for T21, T18 and T13 in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
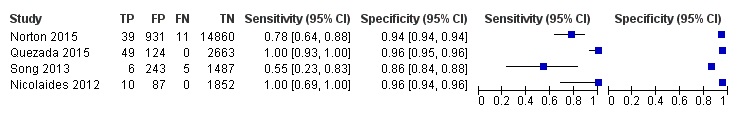
Forest plot of traditional screening tests for autosomes (T21, T18 and T13 combined) in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, TN: true negative and TP: true positive.
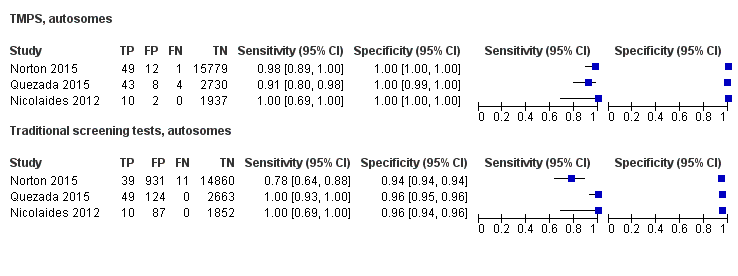
Forest plot of comparative studies of TMPS and traditional screening tests for autosomes (T21, T18 and T13 combined) in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, TN: true negative and TP: true positive.

Forest plot of traditional screening tests for autosomes (T21, T18 and T13 combined) in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for autosomes (T21, T18 and T13 combined) in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
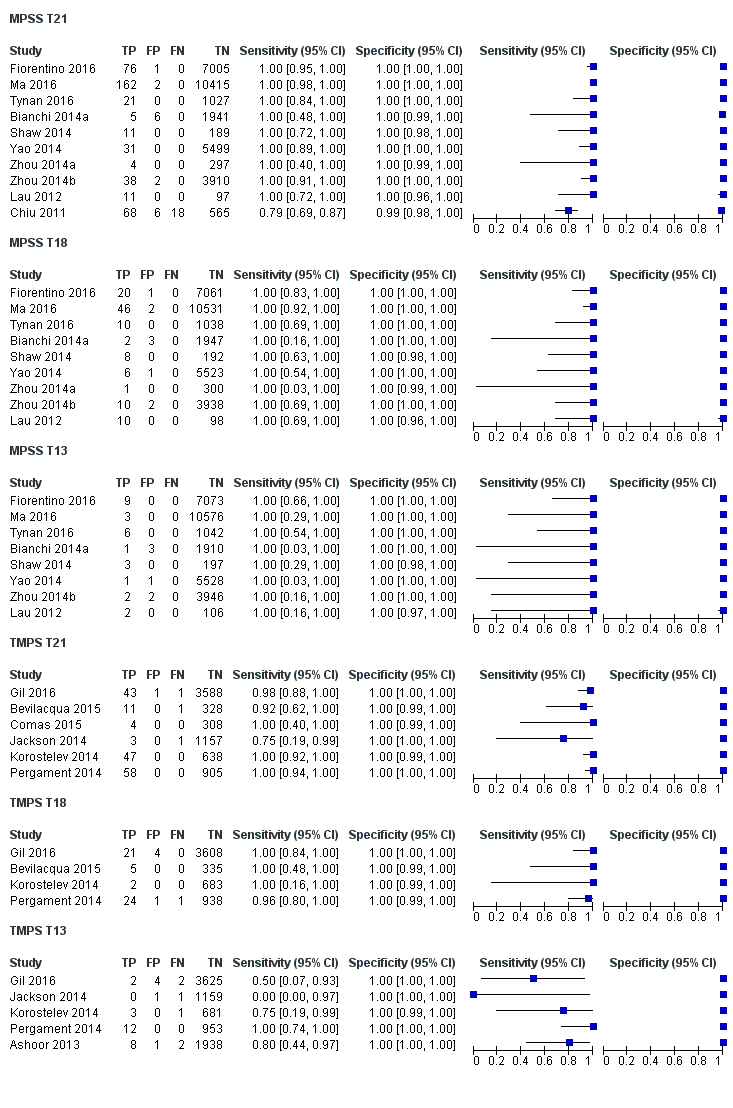
Forest plot of MPSS and TMPS for T21, T18 or T13 in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
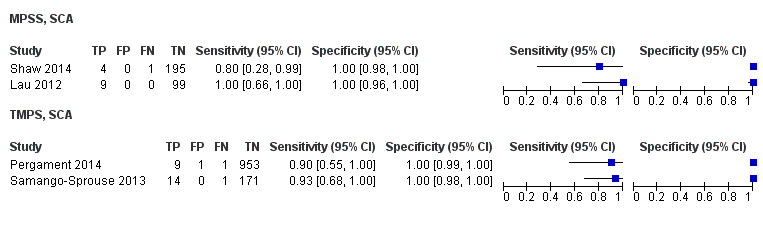
Forest plot of MPSS and TMPS for SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined) in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Forest plot of MPSS and TMPS for 45,X, 47,XXY or 47,XYY in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.

Traditional screening tests, autosomes.

Traditional screening tests T21.

Traditional screening tests T18.

Traditional screening tests T13.
| Summary characteristics of included studies | |
| Review question | What is the diagnostic accuracy of massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS) using circulating cell‐free DNA (ccfDNA) in maternal blood for the detection of common fetal aneuploidies (T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY) in pregnant women according to their prior risk of fetal aneuploidy? |
| Importance (rationale) | These new genomics‐based non‐invasive prenatal testing (gNIPT) approach report higher sensitivity and lower false positive rate than traditional screening tests. gNIPT is already advertised and marketed. How gNIPT should be used in clinical practice should be assessed in order to provide a framework for its use. |
| Study design | There were 40 prospective cohort studies, 8 retrospective cohort studies, 16 case‐control studies and 1 prospective and retrospective cohort study. |
| Population | Pregnant women of any age, ethnicity and gestational age, with singleton or multifetal pregnancy who had a screening test for fetal aneuploidy using gNIPT and received a reference standard. 42 studies enrolled pregnant women selected at high risk of fetal aneuploidy, 5 enrolled unselected pregnant women undergoing aneuploidy screening and 18 enrolled pregnant women from a mixed‐risk population of fetal aneuploidy. 48 studies included only women with singleton pregnancy, 5 included only multifetal pregnancies, 4 included either type of pregnancy and 8 did not report type of pregnancy. 10 studies included only women in the first trimester (15 weeks or less), 21 studies included women in the first 2 trimesters (29 weeks or less), 24 studies included women in the 3 trimesters (42 weeks or less) and 10 studies (15%) did not report gestational age. |
| Index tests | gNIPT by MPSS (44 studies) or TMPS (21 studies), including 5 studies that compared a gNIPT with a traditional screening test. 37 studies were industry‐funded or were written by 1 or more authors affiliated with a company who sells gNIPT. 22 studies were not reported to be funded by industry but samples were sequenced and analysed by a commercial laboratory. 3 studies had no links with industry. |
| Target conditions | 36 studies reported results for only autosomes (T21, T18, T13), 4 for only SCA (45,X, 47,XXY, 47,XXX and 47,XYY), and 25 for both autosomes and SCA. |
| Reference standard | Fetal karyotyping performed on cells obtained from chorionic villi sampling, amniotic fluid, placental tissue, a fetus lost by miscarriage or other equivalent and recognised methods on the same materials for autosomes and SCA. If fetal karyotyping was not performed, we used neonatal clinical examination or medical records from birth (for autosomes only). Only 1 reference standard was used for all pregnant women included in 36 studies while multiple reference standards were used in 29 studies. |
| Risk of bias | The QUality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool was used to assess the methodological quality of included studies. No study was assessed as being at low risk of bias across all domains. For the patient selection domain, no study was assessed as being at low risk of bias. For the index test, reference standard and flow and timing domains, the risk of bias was low for 94%, 77% and 23% of studies, respectively. |
| Applicability concerns | Applicability was of low concern for all studies in the index test and reference standard domains because the studies matched the review question. In the patient selection domain, 47 (71%) studies were judged to be of low applicability concern because they included pregnant women matching the review question. |
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13. | |
| Performance of gNIPT for detection of T21 | |||||||
| Test strategy | Number of studies | Number of affected pregnancies (Number of unaffected pregnancies)a | Sensitivity % (95% CI) | Specificity % (95% CI) | Median prevalenceb % (range) | Missed cases (FN)c | False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 8 (1733) | 100 (67.6 to 100) | 100 (99.8 to 100) | 0.46 (0.24 to 5.21) | 0 | 0 |
| TMPS | 4 | 88 (20,679) | 99.2 (78.2 to 100) | 100 (> 99.9 to 100) | 4 | 0 | |
| Traditional screening teste | 1 | 38 (15,803) | 78.9 (63.7 to 88.9) | 94.6 (94.2 to 94.9) | 97 | 5375 | |
| Implications |
| ||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 30 | 1048 (15,937) | 99.7 (98.0 to 100) | 99.9 (99.8 to 100) | 4.95 (0.44 to 27.66) | 15 | 95 |
| TMPS | 6 | 246 (4380) | 99.2 (96.8 to 99.8) | 100 (99.8 to 100) | 40 | 0 | |
| Difference between MPSS and TMPS | 0.53 (‐0.73 to 1.78) | ‐0.03 (‐0.11 to 0.04) | NA | ||||
| Implications |
| ||||||
| MPSS: massively parallel shotgun sequencing, NA; not applicable, TMPS: targeted massively parallel sequencing, T21: trisomy 21. aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered. cMissed cases per 100,000 tested. FN: false negatives. dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman. eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test. | |||||||
| Performance of gNIPT for detection of T18 | |||||||
| Test strategy | Number of studies | Number of affected pregnancies (Number of unaffected pregnancies)a | Sensitivity % (95% CI) | Specificity % (95% CI) | Median prevalenceb % (range) | Missed cases (FN)c | False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 2 (1739) | 100 (34.3 to 100) | 99.9 (99.7 to 100) | 0.11 (0.06 to 0.36) | 0 | 100 |
| TMPS | 3 | 22 (20,553) | 90.9 (70.0 to 97.7) | 100 (99.9 to 100) | 10 | 0 | |
| Traditional screening teste | 1 | 10 (15,831) | 80.0 (49.0 to 94.3) | 99.7 (99.6 to 99.8) | 22 | 300 | |
| Implications |
| ||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 28 | 332 (16,180) | 97.8 (92.5 to 99.4) | 99.9 (99.8 to 100) | 1.46 (0.22 to 17.02) | 32 | 99 |
| TMPS | 5 | 112 (4010) | 98.2 (93.1 to 99.6) | 100 (99.8 to 100) | 26 | 0 | |
| Difference between MPSS and TMPS | ‐0.41 (‐4.11 to 3.28) | ‐0.06 (‐0.14 to 0.03) | NA | ||||
| Implications |
| ||||||
| MPSS: massively parallel shotgun sequencing, NA: not applicable, TMPS: targeted massively parallel sequencing, T18: trisomy 18. aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered. cMissed cases per 100,000 tested. FN: false negatives. dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman. eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test. | |||||||
| Performance of gNIPT for detection of T13 | |||||||
| Test strategy | Number of studies | Number of affected pregnancies (Number of unaffected pregnancies)a | Sensitivity % (95% CI) | Specificity % (95% CI) | Median prevalenceb % (range) | Missed cases (FN)c | False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 1 (1740) | 100 (20.7 to 100) | 100 (99.8 to 100) | 0. 12 (0.01 to 0.52) | 0 | 0 |
| TMPS | 3 | 8 (14,154) | 65.1 (9.16 to 97.2) | 100 (99.9 to 100) | 41 | 0 | |
| Traditional screening teste | 1 | 2 (11,183) | 50.0 (9.45 to 90.5) | 99.7 (99.6 to 99.8) | 59 | 300 | |
| Implications |
| ||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 20 | 128 (13,810) | 95.8 (86.1 to 98.9) | 99.8 (99.8 to 99.9) | 1.09 (0.04 to 3.54) | 46 | 198 |
| TMPS | 2 | 20 (293) | 100 (83.9 to 100)f | 100 (98.7 to 100)f | 0 | 0 | |
| Implications |
| ||||||
| MPSS: massively parallel shotgun sequencing, NA: not applicable, TMPS: targeted massively parallel sequencing, T13: trisomy 13. aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered. cMissed cases per 100,000 tested. FN: false negatives. dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman. eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test. fSimple pooling used to obtain summary estimates of sensitivity, specificity or both. | |||||||
| Performance of gNIPT for detection of 45,X | |||||||
| Test strategy | Number of studies | Number of affected pregnancies (Number of unaffected pregnancies)a | Sensitivity % (95% CI) | Specificity % (95% CI) | Median prevalenceb % (range) | Missed cases (FN)c | False positives (FP)d |
| Selected high‐risk pregnant women | |||||||
| MPSS | 12 | 119 (7440) | 91.7 (78.3 to 97.1) | 99.6 (98.9 to 99.8) | 1.04 (0.27 to 18.58) | 86 | 396 |
| TMPS | 4 | 79 (985) | 92.4 (84.1 to 96.5) | 99.8 (98.3 to 100) | 79 | 198 | |
| Difference between MPSS and TMPS | ‐0.74 (‐11.1 to 9.60) | ‐0.23 (‐0.82 to 0.36) | NA | ||||
| Implications |
| ||||||
| 45,X: Turner syndrome, MPSS: massively parallel shotgun sequencing, NA: not applicable, TMPS: targeted massively parallel sequencing. aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered. cMissed cases per 100,000 tested. FN: false negatives. dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman. | |||||||
| Performance of gNIPT for detection of autosomes aneuploidies (T21, T18 and T13 combined) | |||||||
| Test strategy | Number of studies | Number of affected pregnancies (Number of unaffected pregnancies)a | Sensitivity % (95% CI) | Specificity % (95% CI) | Median prevalenceb % (range) | Missed cases (FN)c | False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 11 (1730) | 100 (74.1 to 100) | 99.9 (99.7 to 100) | 0,63 (0.32 to 5.73) | 0 | 99 |
| TMPS | 4 | 118 (20,649) | 94.9 (89.1 to 97.7) | 99.9 (99.8 to 99.9) | 32 | 99 | |
| Traditional screening teste | 4 | 120 (22,247) | NDf | ND | |||
| Implications |
| ||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 32 | 1508 (15,797) | 98.8 (97.2 to 99.5) | 99.9 (99.7 to 100) | 5.85 (0.67 to 46.81) | 70 | 94 |
| TMPS | 7 | 378 (4282) | 98.9 (97.2 to 99.6) | 99.9 (99.8 to 100) | 64 | 94 | |
| Difference between MPSS and TMPS | ‐0.11 (‐1.58 to 1.35) | ‐0.08 (‐0.22 to 0.07) | NA | ||||
| Implications |
| ||||||
| MPSS: massively parallel shotgun sequencing, NA: not applicable, ND: no data available, TMPS: targeted massively parallel sequencing, T13: trisomy 13, T18: trisomy 18, T21: trisomy 21. aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered. cMissed cases per 100,000 tested. FN: false negatives. dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman. eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test. fSummary sensitivity and specificity were not obtained for traditional screening tests because the four studies used different cut‐offs to determine test positivity. Three of the four studies compared TMPS and traditional screening tests in the same population (direct comparison). | |||||||
| Performance of gNIPT for detection of sex chromosome aneuploidies (45,X, 47,XXX, 47,XXY and 47,XYY combined) | |||||||
| Test strategy | Number of studies | Number of affected pregnancies (Number of unaffected pregnancies)b | Sensitivity % (95% CI) | Specificity % (95% CI) | Median prevalencec % (range) | Missed cases (FN)d | False positives (FP)e |
| Selected high‐risk pregnant women | |||||||
| MPSS | 12 | 151 (7452) | 91.9 (73.8 to 97.9) | 99.5 (98.8 to 99.8) | 1.53 (0.45 to 18.58) | 124 | 492 |
| TMPS | 4 | 96 (968) | 93.8 (86.8 to 97.2) | 99.6 (98.1 to 99.9) | 95 | 394 | |
| Difference between MPSS and TMPS | ‐1.85 (‐13.3 to 9.60) | ‐0.06 (‐0.82 to 0.71) | NA | ||||
| Implications |
| ||||||
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, MPSS: massively parallel shotgun sequencing, NA: not applicable, ND: no data available, TMPS: targeted massively parallel sequencing aWe did not assess the accuracy of gNIPT individually for 47,XXX, 47,XXY and 47,XYY due to paucity data. bUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. cThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered. dMissed cases per 100,000 tested. FN: false negatives. eFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman. | |||||||
| Target condition | Affected birthsa /100,000 | Clinical features | Prognosis |
| T21 | 140 to 230b,c | Intellectual disability (mild to moderate), neurodevelopmental problems, characteristic dysmorphic features, congenital defects (cardiac (44% to 58%) and gastrointestinal system (4% to 10%)), vision or hearing impairment (38% to 80%) and obstructive sleep apnoea syndrome (57%)d,e | Mean and median life expectancies are estimated to be 51 and 58 years oldf |
| T18 | 59c | Severe intellectual disability and a wide range of significant malformations (cardiac defects, gastrointestinal system defects, renal anomalies, central nervous system defects (apnoea and seizures))d,g | Most affected fetuses die in utero. Median survival has been estimated at 14 days (95% confidence interval (CI) 10 to 20) and 8% (95% CI 4 to 14) reach 1 year of ageh |
| T13 | 23c | Severe intellectual disability, seizures and several dysmorphic features, malformations of the extremities, cardiac defects, renal anomalies, and abdominal wall defectsd,i | Most affected fetuses die in utero. Median survival time has been estimated at 10 days (95% CI 7 to 19) and 8% (95% CI 4 to 14) reach 1 year of ageh |
| 45,X | 30 to 50c,j | Learning disabilities (70%), short stature, congenital heart diseases (30%) and gonadal dysgenesis (90% with amenorrhoea and infertility due to early ovarian failure)k,l | Mortality in 45,X women is 3‐fold higher than in the general population with an average life span of 69 yearsm |
| 47,XXY | 12c | Learning disabilities (> 75%), small testes (> 95%), azoospermia (> 95%), male infertility (91% to 99%), decreased testosterone level (63% to 85%) and gynaecomastia (38% to 75%)l,n | Life expectancy is slightly shorter (approximately 2 years) than euploid menn |
| 47,XXX | 6c | Developmental delays (motor and speech), learning or intellectual disability, attention deficits (25% to 35%), mood disorders (anxiety and depression), tall stature (80% to 89%), clinodactyly (42% to 65%), hypotonia in infancy (55% to 71%), genitourinary malformations and congenital heart defectso | Mortality significantly increased with a median survival age of 70.9 years compare to 81.7 years for euploid femalesp |
| 47,XYY | 3c | Developmental delays (speech, language and motor), attention deficit disorder (52%), tall stature (78%), central adiposity, macrocephaly (33%), hypotonia (63%), clinodactyly (52%), hypertelorism (59%) and testicular enlargement for age (50%) but no increase in genital anomaliesq | Mortality increased with a reduction of life span of 10.3 years compared to euploid menr |
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13. aIncluding live births, fetal deaths and terminations of pregnancy. b(Christianson 2006; Parker 2010) e(Irving 2012; Weijerman 2010) f(Wu 2013b) g(Cereda 2012) h(Wu 2013a) i(Chen 2009) k(Karnis 2012; Mazzanti 1998; Sybert 2004) l(Tyler 2004) m(Saenger 1996; Schoemaker 2008) n(Groth 2013) q(Bardsley 2013; Leggett 2010) r(Stochholm 2010a). | |||
| Test name (Company, country) | Method | Aneuploidy | Reported sensitivity % (95% CI) | Reported specificity % (95% CI) | Reported false positive rate % |
| Bambni™ Test (Berry Genomics Co. Ltd, China) | MPSS | T21 | 100.0 (ND) | > 99.9 (ND) | < 0.1 |
| T18 | 100.0 (ND) | > 99.9 (ND) | < 0.1 | ||
| T13 | 100.0 (ND) | > 99.9 (ND) | < 0.1 | ||
| 45,X | 100.0 (ND) | 99.8 (ND) | 0.0 | ||
| 47,XXX | 100.0 (ND) | 100.0 (ND) | 0.1 | ||
| 47,XXY | 100.0 (ND) | 100.0 (ND) | 0.0 | ||
| 47,XYY | 100.0 (ND) | 100.0 (ND) | 0.0 | ||
| GENOMOM (Genome Care, Korea) | MPSS | T21, T18 and T13 | 99.0 (ND) | ND | ND |
| SCA | 95.0 (ND) | ND | ND | ||
| Harmony™ prenatal test (Ariosa Diagnostics, Inc., USA) | Oligo TMPS | T21 | > 99.0 (ND) | > 99.9 (ND) | < 0.1 |
| T18 | 97.4 (ND) | > 99.9 (ND) | < 0.1 | ||
| T13 | 93.8 (ND) | > 99.9 (ND) | < 0.1 | ||
| 45,Xb | 96.3 (81.7 to 99.8) | 99.5 (98.1 to 99.9) | 0.5 | ||
| 47,XXXb | 100.0 (ND) | 99.5 (98.1 to 99.9) | 0.5 | ||
| 47,XXYb | 100.0 (61.0 to 100.0) | 100.0 (99.0 to 100.0) | 0.0 | ||
| IONA® test (Premaitha Health plc, UK) | MPSS | T21 | > 99.0 (ND) | > 99.0 (ND) | < 1.0 |
| T18 | > 99.0 (ND) | > 99.0 (ND) | < 1.0 | ||
| T13 | > 99.0 (ND) | > 99.0 (ND) | < 1.0 | ||
| (Laboratoire CERBA, France) | MPSS | T21, T18 and T13 | > 99.8 (ND) | > 99.8 (ND) | < 0.2 |
| MaterniT21™ Plus test (Sequenom Inc., USA) | MPSS | T21 | 99.1 (96.6 to 99.9) | 99.9 (99.7 to 99.9) | 0.1 |
| T18 | > 99.9 (93.9 to 100.0) | 99.6 (99.3 to 99.7) | 0.4 | ||
| T13 | 91.7 (61.0 to 99.0) | 99.7 (98.5 to 99.5) | 0.3 | ||
| combined sex aneuploidies | 96.2 (ND) | 99.7 (ND) | 0.3 | ||
| MomGuard™ (LabGenomics, Korea) | MPSS | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY | > 99.0 (ND) | ND | ND |
| NIFTY™ test (Bejing Genomics Institute (BGI), China) | MPSS | T21 | 99.2 (ND) | 100 (ND) | 0 |
| T18 | 98.2 (ND) | 100 (ND) | 0 | ||
| T13 | 100 (ND) | 100 (ND) | 0 | ||
| 45,X | > 99.9 (ND) | > 99.9 (ND) | < 0.1 | ||
| Panorama™ prenatal testc (Natera, Inc., USA) | SNP TMPS | T21 | > 99.9 (ND) | 100 (ND) | 0 |
| T18 | > 96.4 (ND) | > 99.9 (ND) | < 0.1 | ||
| T13 | > 99.9 (ND) | 100 (ND) | 0 | ||
| 45,X | > 92.9 (ND) | > 99.9 (ND) | < 0.1 | ||
| PrenaTest® (LifeCodexx AG, Germany) | MPSS | T21 | 98.7 (ND) | 99.9 (ND) | 0.1 |
| T18 | 100 (ND) | ||||
| T13 | 100 (ND) | ||||
| 45,X | 90.9 (ND) | 98.8 (ND) | 1.2 | ||
| 47,XYY | 100 (ND) | ||||
| Prendia (Genesupport, Switzerland) | MPSS | T21 | 100.0 (88.8 to 100.0) | 100.0 (98.0 to 100.0) | 0.0 |
| T18 | 95.8 (76.8 to 99.7) | 100.0 (97.0 to 100.0) | 0.0 | ||
| T13 | 100.0 (74.6 to 100.0) | 100.0 (98.1 to 100.0) | 0.0 | ||
| 45,X | 100.0 (74.6 to 100.0) | 100.0 (98.1 to 100.0) | 0.0 | ||
| 47,XXX | 100.0 (46.2 to 100.0) | 100.0 (98.2 to 100.0) | 0.0 | ||
| Tranquility (Genoma, Switzerland) | MPSS | T21 | 99.9 (ND) | 99.8 (ND) | 0.2 |
| T18 | 99.9 (ND) | 99.9 (ND) | 0.1 | ||
| T13 | 99.9 (ND) | 99.7 (ND) | 0.3 | ||
| verifi® prenatal test (Illumina, Inc., USA) | MPSS | T21 | 99.5 (98.7 to 99.5) | 99.8 (98.9 to 99.9) | 0.2 |
| T18 | 97.3 (94.2 to 98.2) | 99.7 (99.5 to 99.9) | 0.3 | ||
| T13 | 98.0 (95.6 to 98.9) | 99.8 (99.8 to 99.9) | 0.2 | ||
| 45,X | 95.0 (75.1 to 99.9) | 99.0 (97.6 to 99.7) | 1.0 | ||
| VisibiliT™ (Sequenom Inc., USA) | MPSS | T21 | > 99.0 (80.8 to 100) | > 99.9 (99.5 to 100) | < 0.1 |
| T18 | > 99.0 (65.5 to 100) | > 99.9 (99.5 to 100) | < 0.1 | ||
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13 CI: confidence interval, MPSS: massively parallel shotgun sequencing, ND: no data available, TMPS: targeted massively parallel sequencing and SNP: single nucleotide polymorphism. a(Ariosa Diagnostics 2016; BGI 2014; BGI 2016; Berry Genomics 2016; Genoma 2016; Genome Care 2016; Illumina 2014; Illumina 2016; LabGenomics 2016; LifeCodexx 2016; Natera 2016; Genesupport 2016; Premaitha Health plc 2016; Sequenom 2016). b(Hooks 2014). cDNA of maternal and paternal origin are needed. | |||||
| Screening tests | First trimester (before 14 weeks’ gestation) | Second trimester (14 to 20 weeks’ gestation) |
| Ultrasonography |
|
|
| Combined test |
| NA |
| Triple test | NA |
|
| Quadruple test | NA |
|
| Sequential testb |
|
|
| Contingent testb |
|
|
| Serum integrated testc |
|
|
| Integrated testc |
|
|
| Maternal age is often included in the algorithm for prenatal screening tests. AFP: alpha‐fetoprotein, hCG: human chorionic gonadotropin, NA: not applicable, NT: nuchal translucency, PAPP‐A: pregnancy associated plasma protein A and uE3: unconjugated estriol. a(Gekas 2009; Okun 2008; Wald 2005). | ||
| Study ID | Target condition(s) | Study design and participants | Prior risk | Index test details | Cutpoint | Reference standard | Comparator |
| MPSS | |||||||
| T21 |
| High risk |
| Z score of 3 | Fetal karyotypea | ||
| T21, T18, T13 |
| High risk |
| Z score of 3 for T21; 3.95 for T18 and T13 | Fetal karyotype or neonatal clinical examination | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13, 45,X |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13 |
| High, low and without prior risk |
| NCV of 4; resequenced if NCV is between 3 and 4 | Fetal or postnatal karyotype, neonatal clinical examination or medical record from birth | Standard screening (T21 only with mixed cutpoints) which include first‐trimester combined test or a second‐trimester result (quadruple, serum integrated, fully integrated, or sequential). | |
| T21, T18, T13 |
| High risk |
| NR | Fetal karyotype | ||
| T21, T18, T13 |
| High risk |
| Z score of 3 | Fetal karyotype | ||
| T18, T13 |
| High risk |
| Z score of 3 | Fetal karyotype | ||
| T21 |
| Mostly high (> 1/300) and some intermediate risk (between 1/300 and 1/1000) |
| Z score of 3 | Fetal karyotype | ||
| T21 |
| High risk |
| Z score of 2.5 | Fetal karyotype | ||
| T21, T18, T13 |
| Mostly high risk and without prior risk |
| NCV of 4; aneuploidy suspected if NCV is between 3 and 4 | Fetal karyotype or neonatal clinical examination | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| NR | Fetal karyotype | ||
| T21, T18 |
| High risk |
| L score of 1 and t score of 2.5 including warning zone | Fetal karyotype | ||
| T21, T18 |
| High risk |
| Z score of 2.566 for T21; 2.459 for T18. | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47,XXY, 47, XYY |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13 |
| High risk |
| Z score of 4 (unclassified if Z score is between 3 and 4) and WISECONDOR of 1% | Fetal karyotype | ||
| T21, T18, T13 |
| High risk |
| T score of 3 | Fetal karyotype or newborn outcome | ||
| T21 |
| High risk |
| Z score of 2.10 for Ion Proton™ | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| Mostly high risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13 and SCA (no case found) |
| High risk |
| Z score of 4 (intermediate risk if Z score is between 2.5 and 4) for T21 and T18; 2.8 for T13 (intermediate risk if Z score is between 1.9 and 2.8) | Fetal or neonatal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Z score of 3 | Fetal karyotype | ||
| T21, T18, T13 |
| High and low risk |
| Z score of 3 | Fetal karyotype or postnatal follow‐up | ||
| 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Different cutpoints used for the four SCAb | Fetal karyotype | ||
| T21, T18, T13 |
| High risk |
| Z score of 3 for T21; 3.88 for T18; 7.17 for T13 | Fetal karyotype | ||
| T21, T18, T13 |
| High risk |
| Likelihood ratio of 1 and maternal age‐adjusted probability risk score | Fetal karyotype or medical record from birth | ||
| T21, T18, T13 |
| High risk |
| Likelihood ratio of 1 and maternal age‐adjusted probability risk score | Fetal karyotype or medical record from birth | ||
| T21, T18, T13 |
| High risk |
| NR (authors used the same gNIPT than Papageorghiou 2016a) | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype or medical record from birth | ||
| T21, T18, T13, 45,X |
| High risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47, XXX, 47,XXY, 47,XYY |
| High and low risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype or medical record from birth | ||
| T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY (SCA data not shown in this review) |
| Without prior risk |
| Z score of 3 | Fetal or postnatal karyotype or medical record from birth | Triple test for T21 and T18 (cutpoint of 1 in 270). | |
| T21, T18, T13, 45,X, 47,XXX, 47,XYY |
| High risk |
| Z score of 3 | Fetal karyotype or neonatal clinical examination or both | ||
| T21, T18, T13 |
| High risk |
| MAD‐based Z score of 3 for T21; 3.2 for T18; 3.9 for T13 | Fetal karyotype | ||
| T21, T18, T13, 45,X |
| High risk |
| T score of 5 for T21 and T18; 4 for T13; 0.04 Chrom. X and 0.04 Chrom. Y for 45,X | Fetal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| L score of 1 and t score of 2.5 | Fetal karyotype or medical record from birth | ||
| T21, T18, T13 |
| High and without prior risk |
| risk score of 1% | Fetal karyotype or medical record from birth | ||
| T21, T18, T13, 45,X |
| High risk |
| NR | Fetal or neonatal karyotype or clinical examination at 42 days after birth or both | ||
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Z score of 3 for T21, T18 and T13; ‐3 for Chrom. X and 3 for Chrom. Y for sex Chrom. classification. | Fetal karyotype or clinical follow‐up to 6 months from birth | ||
| T21, T18, T13 and SCA (SCA data not shown in this review) |
| High, low and without prior risk |
| Different cutpoints used for autosomes and SCAb | Fetal karyotype or clinical follow‐up | ||
| T21, T18, 45,X, 47,XXX (SCA data not shown in this review) |
| High risk |
| Z score of 3 for T21 (no other cutpoint reported) | Fetal or neonatal karyotype or neonatal clinical examination | ||
| T21, T18, T13 |
| High, low and without prior risk |
| L score of 1 and t score of 2.5 | Fetal or neonatal karyotype or birth outcome | ||
| T21, T18, T13 |
| High, low and without prior risk |
| L score of 1 and t score of 2.5 | Fetal or neonatal karyotype or birth outcome | ||
| TMPS | |||||||
| T21, T18 |
| High risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype | ||
| T13 |
| High and low risk |
| FORTE risk score of 1% | Fetal karyotype or neonatal clinical examination | ||
| T21, T18, T13 |
| High and without prior risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or neonatal karyotype | ||
| T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY (SCA data not shown in this review) |
| High and without prior risk |
| Harmony™ prenatal test: NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype or neonatal clinical examination | ||
| T21, T18, T13 |
| Without prior risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype | ||
| T21, T18, T13 |
| High and intermediate riskc |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or postnatal karyotype or neonatal clinical examination | ||
| T13 |
| High risk |
| NR | Fetal karyotype or genetic testing of cord blood, buccal, saliva or products of conception | ||
| 45,X, 47,XXX, 47, XXY, 47,XYY |
| High risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype | ||
| T21, T18, T13 |
| High and low risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype or medical record from birth | ||
| T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY |
| High and without prior risk |
| NR | Fetal karyotype or medical record from birth | ||
| T21, T18 |
| Without prior risk |
| Risk score of 1% | Fetal karyotype or neonatal clinical examination | First‐trimester combined test (cutpoint of 1 in 150). | |
| T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| NR | Fetal karyotype | ||
| 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| FORTE risk score of 1% | Fetal karyotype | ||
| T21, T18 |
| High risk |
| FORTE risk score of 1% | Fetal karyotype | ||
| T21, T18, T13 |
| Without prior risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or postnatal karyotype, neonatal clinical examination or medical record from birth | First‐trimester combined test (cutpoint of 1 in 270 for T21 and 1 in 150 for T18 and T13). | |
| T21, T18, T13, 45,X |
| High and low risk |
| NR | Fetal karyotype or genetic testing of cord blood, buccal, saliva or products of conception or birth outcome | ||
| T21, T18, 45,X, 47,XXX, 47,XXY, 47,XYY |
| High risk |
| Risk score of 1% | Fetal karyotype | ||
| T21, T18, T13 |
| Without prior risk |
| NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or postnatal karyotype, neonatal clinical examination or medical record from birth | First‐trimester combined test (cutpoint of 1 in 100 for T21). | |
| 45,X, 47,XXX, 47,XXY, 47,XYY |
| High and low risk |
| NR | Fetal karyotype or genetic testing of cord blood, buccal, saliva or products of conception | ||
| T21, T18 |
| High risk |
| NR | Fetal karyotype | ||
| T21 |
| High risk |
| FORTE risk score of 1% | Fetal karyotype | ||
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, DANSR: digital analysis of selected regions, FF: fetal fraction DNA, FORTE: fetal‐fraction optimised risk of trisomy evaluation, MAD: Median absolute deviation, MPSS: massively parallel shotgun sequencing, NATUS: Next‐generation Aneuploidy Test Using SNPs, NCV: normalised chromosome value, SCA: sex chromosome aneuploidy, SNP: single‐nucleotide polymorphism,TMPS: targeted massively parallel sequencing, T21: trisomy 21, T18: trisomy 18 and T13: trisomy 13. aFetal karyotype include traditional banding techniques, spectral karyotype, fluorescence in situ hybridisation, array comparative genomic hybridisation or quantitative fluorescence polymerase chain reaction. bDifferent cutpoints used for autosomes or SCA as follows: Bianchi 2012: NCV of 4 (aneuploidy suspected if NCV is between 2.5 and 4) for T21, T18, and T13; NCV for Chrom. X of ‐4 and NCV for Chrom. Y of 2.5 for 45,X; NCV for Chrom. X of 4 and NCV for Chrom. Y of 2.5 for 47,XXX; NCV for Chrom. X between ‐2.5 and 2.5 and NCV for Chrom. Y > 33 for 47,XXY; NCV for Chrom. X of ‐4 and NCV for Chrom. Y of 4 for 47,XYY with NCV for Chrom. Y is two times greater than expected NCV Chrom. X. Bianchi 2013: NCV of 4 (aneuploidy suspected if NCV is between 3 and 4) for T21, T18, and T13; NCV for Chrom. X of ‐3 and NCV for Chrom. Y of 3 for 45,X. Jiang 2012: t score of 3 and logarithmic LR of 1 for T21, T18 and T13; if female fetus, t score of ‐2.5 for 45,X and 47,XXX; t score of 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X and Y independently for 47,XXY and 47,XYY. Lau 2012: Z score of 3 for T21, T18 and T13; if female fetus, Z score for Chrom. X of ‐3 for 45,X; if female fetus, Z score for Chrom. X of 3 for 47,XXX; if male fetus, Z score for Chrom. Y of 3 for 47,XXY. Lefkowitz 2016: Z score of 3 for T21; Z score of 3.95 for T18 and T13; Z scores for SCA see Mazloom 2013. Liang 2013: Z score of 3 for T21; 5.91 for T18; 5.72 for T13; ± 2.91 for Chrom. X and ± 3 for Chrom. Y for sex chromosome classification. Mazloom 2013: Z score of 3.5 for 47,XXX (non‐reportable regions between 2.5 and 3.5); Z score of ‐3.5 for 45,X (non‐reportable regions between ‐2.5 and ‐3.5); Z score of ‐3.5 for 47,XYY with Chrom. Y representation; between ‐3.5 and 3.5 for 47,XXY with Chrom. Y representation. Porreco 2014: Z score of 3 for T21; Z score of 3,95 for T18 and T13; Z score of 3.5 for 47,XXX (non‐reportable regions between 2.5 and 3.5); Z score of ‐3.5 for 45,X (non‐reportable regions between ‐2.5 and ‐3.5); Z score of ‐3.5 for 47,XYY with Chrom. Y representation; Z score between ‐3.5 and 3.5 for 47,XXY with Chrom. Y representation. Sehnert 2011: NCV of 4 (unclassified if NCV is between 2.5 and 4) for T21, T18, and T13; NCV for Chrom. Y of ‐2.0 SDs from the mean of male samples and NCV for Chrom. X of ‐3.0 SDs from the mean of female samples for sex chromosome classification. Shaw 2014: Z score of 3 for T21, T18, and T13; Z score of ‐3 for Chrom. X and 3 for Chrom. Y for sex chromosome classification. Yao 2014: T score of 2.5 for T21, T18 and T13; if female fetus, T score for Chrom. X of ‐2.5 for 45,X and 2.5 for 47,XXX; if male fetus, T score for Chrom. X of 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X (expected value of zero) for 47,XXY; if male fetus, T score for Chrom. X of 2.5 and R‐value (the ratio of the fetal DNA fraction estimated by chromosome Y to that estimated by chromosome X) between 1.8 and 2.2 for 47,XYY. cPregnant women with a first‐trimester combined test selected for their risk of fetal aneuploidy (cutpoint of 1 in 100 for high risk and 1 in 101 to 1 in 2500 for intermediate risk). | |||||||
| Company | Number of studies | Number of affected/unaffected pregnanciesa | Number of studies with pregnant women without prior risk of fetal aneuploidy | Number of studies with high‐risk pregnant women | Number of studies with mixed riskb cohort |
| Ariosa Diagnostics, Inc. | 15 | 594/32,302 | 4 | 6 | 5 |
| Bejing Genomics Institute (BGI) | 12 | 427/24,724 | 0 | 7 | 5 |
| Sequenom, Inc. | 9 | 904/8486 | 0 | 7 | 2 |
| Berry Genomics Co. Ltd | 6 | 147/3414 | 1 | 4 | 1 |
| Natera, Inc. | 6 | 276/2103 | 0 | 3 | 3 |
| Illumina, Inc. | 4 | 273/2342 | 0 | 3 | 1 |
| In‐house | 3 | 114/442 | 0 | 3 | 0 |
| Premaitha Health plc | 3 | 99/579 | 0 | 3 | 0 |
| Genome Care | 2 | 21/235 | 0 | 2 | 0 |
| CERBA | 1 | 113/745 | 0 | 1 | 0 |
| Genoma | 1 | 105/6977 | 0 | 0 | 1 |
| LabGenomics | 1 | 8/84 | 0 | 1 | 0 |
| LifeCodexx AG | 1 | 55/417 | 0 | 1 | 0 |
| Not reported | 1 | 5/148 | 0 | 1 | 0 |
| Total | 65 | 3141/82,998 | 5 | 42 | 18 |
| aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bMixed‐risk cohort included a mix of pregnant women without prior risk, low risk or high risk of fetal aneuploidy. | |||||
| Study ID | Number of pregnant women enrolled | Reasons for exclusion | Number of women with results for 2 x 2 table analysis |
| 976 |
Total: 793 | 183 | |
| 400 |
| 397 | |
| 2167 |
Total: 218 | 1949 | |
| 900 |
Total: 14 | 886 | |
| 2362 |
Total: 2022 | 340 | |
| 2882 |
Total: 2366 In addition, other samples excluded from 2 x 2 tables for censored complex karyotype:
| 503 (T21) 502 (T18) 501 (T13) 489 (45,X) | |
| 2882 |
| 113 | |
| 2052 |
Total for T21 and T18: 100 | 1952 (T21 and T18) 1914 (T13) | |
| 10 |
| 9 | |
| 4664 |
| 27 | |
| 392 |
| 289 | |
| 824 |
Total: 71 (8‐plex) | 753 (8‐plex) | |
| 333 |
Total: 21 | 312 | |
| 207 |
Total: 15 | 192 | |
| 480 |
Total: 31 | 449 | |
| 7103 |
| 7082 | |
| 11,692 |
Total: 8059 | 3633 | |
| > 1000 |
Total: 936 | 64 | |
| 432 |
| 414 | |
| 308 |
| 205 | |
| 189 | NR | 189 | |
| 1228 |
Total: 67 | 1161 | |
| 155 | NR | 155 | |
| 903 | NR | 903 | |
| 375 |
Total: 202 | 173 | |
| 2340 | NR | 2340 | |
| 101 | NR | 101 | |
| 1968 |
Total: 1283 | 685 | |
| 108 | NR | 108 | |
| 93 |
| 92 | |
| 5321 |
Total: 4155 (autosomes) In addition:
Total: 4177 (SCA) | 1166 (autosomes) | |
| 435 |
Total: 23 | 412 | |
| 153 | NR | 153 | |
| 10,598 |
Total: 19 | 10,579 | |
| 1975 |
| 411 | |
| 2230 |
Total: 281 | 1949 | |
| 242 |
| 229 | |
| 177 |
Total: 5 | 172 | |
| 4002 |
Total: 922 | 3080 | |
| 18,955 |
Total: 3114 | 15,841 | |
| 4876 |
Total: 2905 | 1971 | |
| 442 |
Total: 16 | 426 | |
| 442 |
Total: 431 | 11 | |
| 1064 |
Total: 98 In addition,
| 963 (T21) 964 (T18 and 45,X) 965 (T13) | |
| 259 |
Total: 10 | 249 | |
| 242 |
| 241 | |
| 4170 |
In addition,
| 3322 (T21, T18, T13) 3201 (47,XXY, 47,XYY) | |
| 2905 |
Total: 120 | 2785 | |
| 201 |
Total: 15 | 186 | |
| 1014 |
Total: 967 | 47 | |
| 201 |
| 200 | |
| 1916 |
Total: 175 | 1741 | |
| 213 |
Total: 9 | 204 | |
| 338 |
| 167 | |
| 522 |
Total: 50 | 472 | |
| 200 | NR | 200 | |
| 918 |
Total: 17 | 901 | |
| 1100 |
Total: 52 | 1048 | |
| 595 |
Total: 91 | 504 | |
| 136 | NR | 136 | |
| 917 | NR | 917 | |
| 5950 |
| 5530 | |
| 87 | NR | 87 | |
| 306 |
| 301 | |
| 7705 |
Total: 3755 | 3950 | |
| ccfDNA: circulating cell‐free DNA, CVS: chorionic villi sampling, GA: gestational age, gNIPT: genomics‐based non‐invasive prenatal testing, NR: not reported by authors. aSecond time: sample failed the second gNIPT assay. bFirst time: sample failed the initial gNIPT assay. | |||
| Study ID | Failure rate at first attempt (%) | Repeated testsa (%) | Failure rate of repeated tests (%) | Final failure rate total (%) | Aneuploidb samples (%) | Euploidb samples (%) |
| MPSS | ||||||
| 61/244 (25%) | 0 | NA | 61/244 (25%) | NR | NR | |
| 42/892 (4.7%) | 42 (100%) with second aliquot | 6/42 (14.3%) | 6/892 (0.7%) | 2.7% | 0.4% | |
| 16/519 (3.1%) | 0 | NA | 16/356 (3.1%) | NR | NR | |
| 18/1970 (0.9%) | 0c | NA | T21 and T18: 18/1970 (0.9%) T13: 18/1932 (0.9%) | NR | NR | |
| 1/10 (10.0%) | 0 | NA | 1/10 (10.0%) | 50% | 0% | |
| 11/764 (1.4%) | 0 | NA | 11/764 (1.4%) | NR | NR | |
| 20/467 (4.3%) | 20 (100%) resequenced | 18/20 (90%) | 18/467 (3.9%) | NR | NR | |
| 100/7103 (1.4%) | 79 (79%) with new sampling | 0 (0%) | 21/7103 (0.3%) | 0% | 0.3% | |
| NR | 2 with second aliquot or resequenced were in the grey zone (between affected and unaffected) | NR | 11/184 (6%)d | 5.8% | 6.1% | |
| 1/93 (1.1%) | 0 | NA | 1/93 (1.1%) | NR | NR | |
| Autosomes: 42/1208 (3.5%) SCA: 64/1208 (5.3%) | 0 | NA | Autosomes: 42/1208 (3.5%) SCA: 64/1208 (5.3%) | Autosomes: 3.8% SCA: 29.7% | Autosomes: 3.4% SCA: 4.5% | |
| 12/424 (2.8%) | 0 | NA | 12/424 (2.8%) | NR | NR | |
| 5/10,584 (0.05%) | 0 | NA | 5/10,584 (0.05%) | NR | NR | |
| 21/432 (4.9%) | 0 | NA | 21/432 (4.9%) | 11.8% | 4.3% | |
| 110/1988 (5.5%) | 105 (95.5%) with second aliquot and 5 (4.5%) resequenced | 17/110 (15.5%) | 17/1988 (0.9%) | 1.0% | 0.8% | |
| 5/431 (1.2%) | 0 | NA | 5/431 (1.2%) | NR | NR | |
| 1/242 (0.4%) | 0 | NA | 1/242 (0.4%) | 0% | 0.5% | |
| Autosomes: 108/3430 (3.1%) 45,X and 47,XXX: 152/3430 (4.4%) 47,XXY and 47,XYY: 229/3430 (6.7%) | 0 | NA | Autosomes: 108/3430 (3.1%) 45,X and 47,XXX: 152/3430 (4.4%) 47,XXY and 47,XYY: 229/3430 (6.7%) | NR | NR | |
| 73/1814 (4.0%) | 0 | NA | 73/1814 (4.0%) | 0% | 4.0% | |
| 1/205 (0.5%) | 0 | NA | 1/205 (0.5%) | NR | NR | |
| 32/504 (6.3%) | 0 | NA | 32/504 (6.3%) | 3.5% | 6.7% | |
| 21/908 (2.3%) | 16 (76.2%) with new sampling | 2/16 (12.5%) | 7/908 (0.8%) | NR | NR | |
| 52/1100 (4.7%) | 0 | NA | 52/1100 (4.7%) | 0% | 4.9% | |
| 0 | 0 | NA | 0 | NA | NA | |
| 0 | 0 | NA | 0 | NA | NA | |
| 141/3954 (3.6%) | 141 (100%) with new sampling | 4/141 (2.8%) | 4/3954 (0.1%) | NR | NR | |
| Overall range of final assay failure for MPSS | 0% to 25% | 0% to 50% | 0% to 6.7% | |||
| TMPS | ||||||
| 3/400 (0.8%) | 0 | NA | 3/400 (0.8%) | 0% | 1% | |
| 53/2002 (2.6%) | 0 | NA | 53/2002 (2.6%) | 0% | 2.7% | |
| 29/356 (8.1%) | 26 (90%) with 2nd aliquot | 13/26 (50%) | 16/356 (4.5%) | NR | NR | |
| 9/316 (2.8%) | 6 (67%) with new sampling | 1/6 (16.7%) | 4/316 (1.3%) | NR | NR | |
| 15/207 (7.2%) | 0 | NA | 15/207 (7.2%) | 23% | 6% | |
| 99/3698 (2.8%) | 54 (54,5%) with new sampling | 20/54 (37%) | 65/3698 (1.8%) | NR | NR | |
| 4/68 (5.9%) | 0 | NA | 4/68 (5.9%) | 11.8% | 3.9% | |
| 18/432 (4.2%) | 0 | NA | 18/432 (4.2%) | NR | NR | |
| NR | NR | 14 (NR) | 14/1175 (1.2%) | NR | NR | |
| 100/2049 (4.9%) | 0 | NA | 100/2049 (4.9%) | 9.1% | 4.9% | |
| 13/242 (5.4%) | 0 | NA | 13/242 (5.4%) | 6.3% | 5.2% | |
| 5/177 (2.8%) | 0 | NA | 5/177 (2.8%) | 5.1% | 1.7% | |
| 148/3228 (4.6%) | 0 | NA | 148/3228 (4.6%) | NR | NR | |
| 488/16,329 (3.0%) | 0 | NA | 488/16,329 (3.0%) | 20.6% | 2.9% | |
| T21: 88/1051 (8.4%) T18, 45,X: 87/1052 (8.3%) T13: 86/1053 (8.2%) | 0 | NA | T21: 88/1051 (8.4%) T18, 45,X: 87/1052 (8.3%) T13: 86/1053 (8.2%) | All five chromosomes (n = 85): 15.2% | All five chromosomes (n = 85): 7.1% | |
| 10/259 (3.9%) | 0 | NA | 10/259 (3.9%) | 8.4% | 2.1% | |
| 122e/2838 (4.2%) | 110 (90.1%) with new sampling | 41/110 (37.3%) | 53/2838 (1.9%) | 4.1% | 1.8% | |
| 15/201 (7.5%) | 0 | NA | 15/201 (7.5%) | 6.3% | 7.6% | |
| 51/520 (9.8%) | 51 (100%) with 2nd aliquot | 16/51 (31.4%) | 16/520 (3.1%) NR | NR | NR | |
| Overall range of final assay failure for TMPS | 0.8% to 7.5% | 0% to 23% | 1% to 7.63% | |||
| CVS: chorionic villi sampling, FF: fetal fraction DNA, GA: gestational age, NA: not applicable, NR: not reported by authors, QC: quality control. baneuploid: proportion of failed samples of aneuploid cases out of all aneuploid tested with reference standard and gNIPT result. euploid: proportion of failed samples of euploid cases out of all euploid tested with reference standard and gNIPT result. cAuthors decided to resequence 12 samples with gNIPT results. They were in the grey zone (between affected and unaffected) and were resequenced in uniplex. All repeated tests were in affected or unaffected zone. dOnly the final failure rate was reported.The failure rate at first attempt was not reported nor the failure rate of repeated tests. eAuthor reported 123 failed tests but this number included one sample lost in the mail and so did not undergo the sequencing process. | ||||||
| Test | Number of studies | Number of affected pregnancies | Number of unaffected pregnanciesa | |
| 47,XXX | ||||
| Selected high risk pregnant women | MPSS | 5 | 8 | 5441 |
| TMPS | 2 | 6 | 580 | |
| 47,XXY | ||||
| Selected high risk pregnant women | MPSS | 7 | 14 | 6466 |
| TMPS | 3 | 8 | 827 | |
| 47,XYY | ||||
| Selected high risk pregnant women | MPSS | 7 | 11 | 6418 |
| TMPS | 1 | 3 | 169 | |
| aUnaffected pregnancies: we included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unnon affected". | ||||
| Test subgroups | Number of studies | Number of affected pregnancies | Number of unaffected pregnanciesa | Sensitivityb % (95% CI) | Specificityb % (95% CI) | |
| Pregnancy type | ||||||
| Autosomes (T21, T18 and T13 combined), unselected population | ||||||
| MPSS | singleton | 1 | 11 | 1730 | 100 (74.1 to 100) | 99.9 (99.7 to 100) |
| TMPS | singleton | 3 | 107 | 20,468 | 95.5 (87.4 to 98.4) | 99.9 (99.8 to 100) |
| multifetal | 1 | 11 | 181 | 90.9 (62.3 to 98.4) | 100 (97.9 to 100) | |
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | singleton | 19 | 1087 | 11,180 | 98.3 (97.3 to 98.9) | 99.6 (99.5 to 99.7) |
| multifetal | 3 | 21 | 206 | 95.2 (72.9 to 99.3) | 100 (98.2 to 100)c | |
| TMPS | singleton | 7 | 378 | 4282 | 98.9 (97.2 to 99.6) | 99.9 (99.8 to 100) |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | singleton | 7 | 101 | 4690 | 88.3 (52.9 to 98.1) | 99.3 (97.5 to 99.8) |
| TMPS | 4 | 96 | 968 | 93.8 (86.8 to 97.2) | 99.6 (98.1 to 99.9) | |
| Gestational age | ||||||
| Autosomes (T21, T18 and T13 combined), unselected population | ||||||
| MPSS | ≤29 weeks | 1 | 11 | 1730 | 100 (74.1 to 100) | 99.9 (99.7 to 100) |
| TMPS | ≤15 weeks | 4 | 118 | 20,649 | 94.9 (89.1 to 97.7) | 99.9 (99.8 to 99.9) |
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | ≤15 weeks | 3 | 49 | 532 | 100 (92.7 to 100)c | 100 (99.3 to 100)c |
| ≤29 weeks | 12 | 594 | 4605 | 98.3 (96.9 to 99.1) | 99.3 (99.0 to 99.5) | |
| ≤42 weeks | 13 | 729 | 7831 | 98.9 (95.0 to 99.8) | 99.9 (99.8 to 99.9) | |
| TMPS | ≤15 weeks | 2 | 128 | 498 | 99.2 (95.7 to 99.9)c | 100 (99.2 to 100)c |
| ≤29 weeks | 2 | 33 | 535 | 97.0 (84.7 to 99.5)c | 100 (99.3 to 100)c | |
| ≤42 weeks | 2 | 163 | 3084 | 99.4 (95.8 to 99.9) | 99.9 (99.7 to 100) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | ≤15 weeks | 1 | 2 | 202 | 0.00 (0.00 to 65.8) | 99.5 (97.2 to 99.9) |
| ≤29 weeks | 5 | 58 | 996 | 86.5 (63.1 to 96.0) | 95.1 (93.5 to 96.3) | |
| ≤42 weeks | 5 | 89 | 6103 | 95.8 (80.3 to 99.2) | 99.6 (99.4 to 99.7) | |
| TMPS | ≤15 weeks | 2 | 58 | 343 | 93.1 (83.0 to 97.4) | 99.7 (98.0 to 100) |
| ≤42 weeks | 1 | 34 | 380 | 97.1 (85.1 to 99.5) | 98.9 (97.3 to 99.6) | |
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13 CI: confidence interval, MPSS: massively parallel shotgun sequencing, SCA: sex chromosome aneuploidies, TMPS: targeted massively parallel sequencing. aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bFor two or more studies, the sensitivities and specificities are the summary estimates obtained from meta‐analysis. cSimple pooling used to obtain summary estimates of sensitivity, specificity or both. | ||||||
| Study | Sensitivity (true positives/cases) % | Difference % (95% CI) | Specificity (true negatives/unaffecteda) % | Difference % (95% CI) | ||
| MPSS | Traditional screening tests | MPSS | Traditional screening tests | |||
| 100 (11/11) | 54.6 (6/11) | 45.5 (10.0 to 72.0) | 99.9 (1729/1730) | 86.0 (1487/1730) | 14.0 (12.4 to 15.7) | |
| TMPS | Traditional screening tests | TMPS | Traditional screening tests | |||
| 100 (10/10) | 100 (10/10) | 0.00 (‐27.8 to 27.8) | 99.9 (1937/1939) | 95.5 (1852/1939) | 4.38 (3.51 to 5.40) | |
| 98.0 (49/50) | 78.0 (39/50) | 20.0 (7.44 to 33.3) | 99.9 (15,779/15,791) | 94.1 (14,860/15,791) | 5.82 (5.46 to 6.20) | |
| 91.5 (43/47) | 100 (49/49) | ‐8.51 (‐19.9 to 0.40) | 99.7 (2730/2738) | 95.6 (2663/2787) | 4.16 (3.40 to 5.00) | |
| CI: confidence interval, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing. aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. | ||||||
| Test | Number of studies | Number of affected pregnancies | Number of unaffected pregnanciesa | Summary sensitivity % (95% CI) | Summary specificity % (95% CI) | P valueb |
| Case‐control studies excluded | ||||||
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | 22 | 696 | 11,293 | 98.3 (95.1 to 99.4) | 99.9 (99.8 to 100) | 0.72 |
| TMPS | 4 | 219 | 3,813 | 98.6 (95.8 to 99.6) | 99.9 (99.8 to 100) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | 10 | 98 | 5,872 | 91.9 (73.8 to 97.9) | 99.5 (98.8 to 99.8) | 0.41 |
| TMPS | 2 | 6 | 472 | 93.8 (86.8 to 97.2) | 99.6 (98.1 to 99.9) | |
| Exclusion of studies with less than 10 pregnancies with aneuploidy | ||||||
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | 21 | 1458 | 13,921 | 98.7 (96.8 to 99.4) | 99.8 (99.5 to 100) | 0.07 |
| TMPS | 7 | 378 | 4,282 | 98.9 (97.2 to 99.6) | 99.9 (99.8 to 100) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | 6 | 130 | 5,761 | 94.5 (80.6 to 98.6) | 99.4 (97.6 to 99.8) | 0.28 |
| TMPS | 2 | 90 | 496 | 94.4 (87.3 to 97.7) | 99.0 (97.6 to 99.6) | |
| 45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13 CI: confidence interval, MPSS: massively parallel shotgun sequencing, SCA: sex chromosome aneuploidies, TMPS: targeted massively parallel sequencing. aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies. bThe P value indicates the statistical significance of the difference in model fit and was obtained from likelihood ratio tests comparing models with and without a covariate for test type. | ||||||
| Test | No. of studies | No. of participants |
| 1 MPSS T21 Show forest plot | 41 | 50133 |
| 2 MPSS T18 Show forest plot | 38 | 49003 |
| 3 MPSS T13 Show forest plot | 29 | 46090 |
| 4 MPSS 45,X Show forest plot | 14 | 7867 |
| 5 MPSS 47, XXX Show forest plot | 5 | 5449 |
| 6 MPSS 47,XXY Show forest plot | 8 | 6588 |
| 7 MPSS 47,XYY Show forest plot | 8 | 6629 |
| 8 MPSS all 7 aneuploidies Show forest plot | 44 | 50864 |
| 9 MPSS, autosomes Show forest plot | 43 | 50453 |
| 10 MPSS, SCA Show forest plot | 14 | 7911 |
| 11 TMPS T21 Show forest plot | 16 | 32487 |
| 12 TMPS T18 Show forest plot | 12 | 30319 |
| 13 TMPS T13 Show forest plot | 10 | 22868 |
| 14 TMPS 45,X Show forest plot | 6 | 2214 |
| 15 TMPS 47,XXX Show forest plot | 2 | 586 |
| 16 TMPS 47,XXY Show forest plot | 4 | 1021 |
| 17 TMPS 47,XYY Show forest plot | 2 | 358 |
| 18 TMPS all 7 aneuploidies Show forest plot | 21 | 35275 |
| 19 TMPS, autosomes Show forest plot | 18 | 34473 |
| 20 TMPS, SCA Show forest plot | 6 | 2214 |
| 21 Traditional screening tests, autosomes Show forest plot | 5 | 24279 |
| 22 Traditional screening tests T21 Show forest plot | 2 | 17753 |
| 23 Traditional screening tests T18 Show forest plot | 2 | 17747 |
| 24 Traditional screening tests T13 Show forest plot | 1 | 11185 |

