Appendix stump closure during laparoscopic appendectomy
Abstract
This is a protocol for a Cochrane Review (Intervention). The objectives are as follows:
This review will compare all surgical techniques that are being used for appendiceal stump closure during laparoscopic appendectomy.
Background
Appendicitis refers to inflammation of the appendix (Andersen 2011). Appendectomy (surgical removal of the appendix) is performed as an emergency procedure to treat acute appendicitis (Andersen 2005).
Description of the condition
Acute appendicitis, which was first described by Fitz in 1886, is the most common cause of acute abdominal pain (Andersen 2005; Rehman 2011; Wilms 2011). The overall incidence of acute appendicitis varies between 76 and 227 cases per 100,000 population per year in different regions (Addiss 1990;Andreu‐Ballester 2009;Buckius 2011; Lee 2010; Pieper 1982). The overall lifetime risk for acute appendicitis in United States and South Korea is approximately 7%‐8% and 16%, respectively (Addiss 1990;Lee 2010). It affects all age groups with the highest incidence in the second decade (Addiss 1990; Wilms 2011).
The cause of acute appendicitis is an issue of considerable debate (Andersen 2005). Acute appendicitis might be associated with obstruction of the appendix lumen (the inside space of an appendix) which could result in increased intraluminal pressure with transmural tissue necrosis (Andersen 2005). Tissue necrosis is followed by bacterial invasion, leading to inflammation of the appendix (Andersen 2005; Wikipedia 2011).
Description of the intervention
Patients with acute appendicitis usually need appendectomy (irrespective of open or laparoscopic appendectomy) to relieve symptoms and avoid complications. Laparoscopic appendectomy was first described in 1983 (Schier 1998). And then undergoing some modifications(from four ports to three,and then two). In 1992, Pelosi reported the single incision laparoscopic surgery, which had less trauma and was considered as a safe operation technique (Pelosi 1992). Laparoscopic and open appendectomy are both well‐accepted by surgeons, as clinical data have shown that the advantages of laparoscopic appendectomy are clearly provable but not very large. One of the possible drawbacks of the laparoscopic technique is the slightly higher intraabdominal abscess rate (Sauerland 2010). In this context, it was suggested that appendix stump closure techniques play a key role in preventing infectious complications after appendectomy (Krisher 2001).
How the intervention might work
The traditional technique for securing the appendix stump during open appendectomy is the tobacco pursue suture, but this suture is quite difficult to apply during laparoscopic appendectomy (Houben 1998). Therefore, two other techniques have been introduced for laparoscopic appendectomy. The first technique is the Roeder loop, a pre‐tied sliding knot, which was developed by Roeder (a German ENT surgeon ) for tonsillectomy (Röder 1918). After applying two or more of these loops to the base of the appendix, the appendiceal stump can bed in between (Beldi 2006; Shimi 1994). The second technique is the gastrointestinal anastomosis (GIA) stapler (Daniell 1991; Klaiber 1994). By using this device, two rows of small staples are applied to hold tissue edges together, so that automatic dissection can be done between the two rows (Beldi 2006). The GIA stapler can be loaded with different cartridges of staples, thus allowing to suture different types of tissue, such as the appendiceal base and the mesoappendix with its artery. Recently, studies about different types of clips for laparoscopic appendectomy have been described (Hanssen 2007; Partecke 2010; Delibegovic 2009).These clips have the advantages of easy application and low costs.
Why it is important to do this review
Roeder loops and GIA stapling devices are both commonly used during laparoscopic appendectomy. The main difference between the two techniques is currently believed to represent a trade‐off between costs and safety, because the stapler is much more expensive to use but seems to result in a safer closure of the stump. Certainly, the degree of local inflammation and the expertise of the operating surgeon also play a decisive role in the selection of surgical technique. Up to now, there has been no Cochrane review determining the preferred technique for securing the appendiceal stump in laparoscopic appendectomy.
Objectives
This review will compare all surgical techniques that are being used for appendiceal stump closure during laparoscopic appendectomy.
Methods
Criteria for considering studies for this review
Types of studies
We will include randomised controlled trials (RCTs) . We will exclude quasi‐randomized trials (where the allocation is done on the basis of a pseudo‐random sequence, e.g. odd/even hospital number or date of birth, alternation) and non‐randomized studies (Higgins 2011a). All types of publications are eligible for this review regardless of language (English or non‐English) or length (full article or abstract).
Types of participants
We will include patients (irrespective of age, sex, or race) who are about to undergo laparoscopic appendectomy.
Types of interventions
-
Roeder loops versus stapling.
-
Different variants of the Roeder technique (number and positioning of the loops) and the stapler (type and number of staples, positioning of the staple line).
-
Endo‐clips versus Roeder loops.
-
Endo‐clips versus stapling.
Types of outcome measures
Primary outcomes
-
Rate of intraabdominal infections (abscess or peritonitis).
-
Rate of intestinal fistula.
Secondary outcomes
-
Complications.
-
Local bleeding.
-
Stump insufficiency.
-
Ileus
-
Superficial wound infection
-
-
Duration of surgery.
-
Hospital stay.
-
Hospital costs(operation, direct and indirect).
-
Pain/Quality of life.
Search methods for identification of studies
We will design the search strategies with the help of Marija Barbateskovic (Trial Search Coordinator) before searching (irrespective of language, year, or publication status).
Electronic searches
We will search The Cochrane Library; MEDLINE, EMBASE, Science Citation Index Expanded, and China Biological Medicine Database (CBM) during the review preparation (Royle 2003). We have given the preliminary search strategies for the listed databases in Appendix 1, Appendix 2, Appendix 3, and Appendix 4.
Searching other resources
We will search the following databases which include ongoing trials: The World Health Organization International Trials Registry Platform search portal (http://apps.who.int/trialsearch/), ClinicalTrials.gov (http://www.clinicaltrials.gov/), Current Controlled Trials (http://www.controlled‐trials.com/), Chinese Clinical Trial Register (http://www.chictr.org/), and EU Clinical Trials Register (https://www.clinicaltrialsregister.eu/). We will also search the reference lists in relevant publications and meeting abstracts (via http://www.eaes‐eur.org/, http://www.sages.org/ and Conference Proceedings Citation Index) to explore further relevant clinical trials. We also plan to communicate with the authors of RCTs included for more information in the review, if necessary.
Data collection and analysis
We will conduct the systematic review according to the Cochrane Handbook for Systematic Reviews of Intervention (Higgins 2011b) and Colorectal Cancer Group Module (Andersen 2011).
Selection of studies
After completing all searches, we will merge the search results using the software package Endnote X5 (reference management software) and remove duplicate records of the same report. Two independent review authors (Zhou ZG, Cheng Y) will scan the title and abstract of every record identified by the search for inclusion. We will retrieve full text for further assessment if the inclusion criteria are unclear from the abstract. We plan to detect duplicate publication by identifying common authors, centres, details of the interventions, numbers of participants, and baseline data (Higgins 2011c). We intend to correspond with the authors of the RCTs to confirm whether the trial results had been duplicated, if necessary. We will exclude papers not meeting the inclusion criteria and list the reasons for the exclusion. A third review author ( Peng S) will resolve any disagreements between the two authors by discussion, and if required, by consultation with the review group's editors.
Data extraction and management
Two authors (Cheng NS and Liao Y) will independently extract the data, check and enter the data into an electronic data collection form (Microsoft Word) (Figure 1). We will resolve any disagreements between the two authors by consensus.
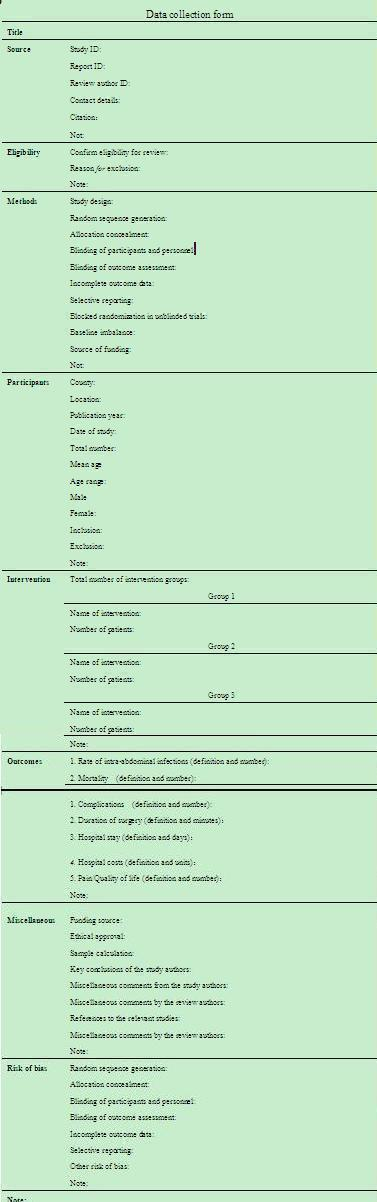
Data collection form (Microsoft Word)
Assessment of risk of bias in included studies
Two review authors (Zhang YL, Zhou J) will assess the methodological quality of the included trails independently. We plan to use the quality checklist recommended by the Cochrane Handbook for Systematic Reviews of Intervention (Table 1) (Higgins 2011d). We will resolve any disagreements by discussion and referral to a third author (Peng S) for adjudication. We intend to present the results of risk of bias by two figures (a 'Risk of bias graph' figure and a 'Risk of bias summary' figure) generated using Review Manager 5 (RevMan 2011).
| Domain | Support for judgement | Review authors’ judgement |
| Random sequence generation. | Describe the method used to generate the allocation sequence in sufficient detail to allow an assessment of whether it should produce comparable groups. | Selection bias (biased allocation to interventions) due to inadequate generation of a randomised sequence: High risk/Low risk/Unclear risk? |
| Allocation concealment. | Describe the method used to conceal the allocation sequence in sufficient detail to determine whether intervention allocations could have been foreseen in advance of, or during, enrolment. | Selection bias (biased allocation to interventions) due to inadequate concealment of allocations prior to assignment: High risk/Low risk/Unclear risk? |
| Blinding of participants and personnel. | Describe all measures used, if any, to blind study participants and personnel from knowledge of which intervention a participant received. Provide any information relating to whether the intended blinding was effective. | Performance bias due to knowledge of the allocated interventions by participants and personnel during the study: High risk/Low risk/Unclear risk? |
| Blinding of outcome assessment. | Describe all measures used, if any, to blind outcome assessors from knowledge of which intervention a participant received. Provide any information relating to whether the intended blinding was effective. | Detection bias due to knowledge of the allocated interventions by outcome assessors: High risk/Low risk/Unclear risk? |
| Incomplete outcome data. | Describe the completeness of outcome data for each main outcome, including attrition and exclusions from the analysis. State whether attrition and exclusions were reported, the numbers in each intervention group (compared with total randomised participants), reasons for attrition/exclusions where reported, and any re‐inclusions in analyses performed by the review authors. | Attrition bias due to amount, nature or handling of incomplete outcome data: High risk/Low risk/Unclear risk? |
| Selective reporting. | State how the possibility of selective outcome reporting was examined by the review authors, and what was found. | Reporting bias due to selective outcome reporting: High risk/Low risk/Unclear risk? |
| Other sources of bias. | State any important concerns about bias not addressed in the other domains in the tool. | Bias due to problems not covered elsewhere in the table: High risk/Low risk/Unclear risk? |
Measures of treatment effect
We will perform the meta‐analyses using the software package Review Manager 5 (RevMan 2011). For dichotomous outcomes, we will calculate the risk ratio (RR) with 95% confidence interval (Deeks 2011). In case of rare events (e.g. mortality), we plan to calculate the Peto odds ratio (Peto OR) (Deeks 2011). For continuous outcomes, we will calculate mean difference (MD) with 95% confidence interval (Deeks 2011). For continuous outcomes with different measurement scales in different randomised clinical trials, we will calculate standardized mean differences (SMD) with 95% confidence interval (Deeks 2011).
Unit of analysis issues
The unit of analysis is each patient. We will analyse data using the generic inverse‐variance method in Review Manager for cluster‐randomized trials (Higgins 2011a). We will combine groups to create a single pair‐wise comparison for trials with multiple intervention groups (Higgins 2011a).
Dealing with missing data
We will contact the original investigators to request further information in case of missing data. If there is no reply, we will perform the analysis on an 'intention‐to‐treat' (ITT) principle, if applicable (Newell 1992). Otherwise, we will use only the available data in the analysis.
Assessment of heterogeneity
We plan to describe the heterogeneity by using the Chi2 test (Deeks 2011). The P value less than 0.10 is considered to be significant heterogeneity (Deeks 2011). We also plan to use the I2 statistic to measure the quantity of heterogeneity. In case of heterogeneity, we will perform the meta‐analysis and interpret the result cautiously. We will explore the clinical heterogeneity by comparing the characteristics of participants, interventions, controls, outcome measures and study designs in the included studies. We plan to undertake the following approaches for explanation and solution: (ⅰ) Check again that the data are correct; (ⅱ) Change the effect measure; (ⅲ) analysis using the random‐effects model; (Ⅳ) sensitivity analysis by excluding potentially biased trails; (ⅴ) subgroup analysis or meta‐regression; (ⅵ) present all trails and provide a narrative discussion (Deeks 2011).
Assessment of reporting biases
If meta‐analysis is possible, we plan to use funnel plots to assess reporting biases (Sterne 2011). Visual asymmetry in funnel plots will be used to determine the reporting biases (Sterne 2011). We will not perform funnel plots if the number of trails included is less than ten (Sterne 2011).
Data synthesis
We will perform the meta‐analyses using Review Manager 5 software provided by The Cochrane Collaboration (RevMan 2011). Two review authors ( Liao Y, Zhou ZG) will check and enter all data into Review Manager independently. We will resolve any disagreements by consensus. For all analyses, we will employ both fixed‐effect and random‐effects models. We will only report the fixed‐effect model results when there is no discrepancy between the two models. In case of discrepancy between the two models, we will report both results.
Subgroup analysis and investigation of heterogeneity
If there is a significant heterogeneity among the RCTs, we plan to perform the following subgroup analyses:
-
Trials with low risk of bias versus trials with high risk of bias.
-
Adults versus children.
-
Complicated(gangrenous or perforated appendicitis) versus uncomplicated.
-
Single incision versus non‐single incision.
-
Male versus female.
Sensitivity analysis
We will perform sensitivity analyses to see whether conclusions are robust to decisions made during the review process following the method as follows:
-
Changing statistics among risk ratio (RR), risk differences (RD) and odds ratios (OR) for dichotomous outcomes.
-
Changing statistics between mean difference (MD) and standardized mean differences (SMD) for continuous outcomes.
-
Excluding trials with high risk of bias.
If the results do not change, they are considered to have low sensitivity. If the results change, they are considered to have high sensitivity.

Data collection form (Microsoft Word)
| Domain | Support for judgement | Review authors’ judgement |
| Random sequence generation. | Describe the method used to generate the allocation sequence in sufficient detail to allow an assessment of whether it should produce comparable groups. | Selection bias (biased allocation to interventions) due to inadequate generation of a randomised sequence: High risk/Low risk/Unclear risk? |
| Allocation concealment. | Describe the method used to conceal the allocation sequence in sufficient detail to determine whether intervention allocations could have been foreseen in advance of, or during, enrolment. | Selection bias (biased allocation to interventions) due to inadequate concealment of allocations prior to assignment: High risk/Low risk/Unclear risk? |
| Blinding of participants and personnel. | Describe all measures used, if any, to blind study participants and personnel from knowledge of which intervention a participant received. Provide any information relating to whether the intended blinding was effective. | Performance bias due to knowledge of the allocated interventions by participants and personnel during the study: High risk/Low risk/Unclear risk? |
| Blinding of outcome assessment. | Describe all measures used, if any, to blind outcome assessors from knowledge of which intervention a participant received. Provide any information relating to whether the intended blinding was effective. | Detection bias due to knowledge of the allocated interventions by outcome assessors: High risk/Low risk/Unclear risk? |
| Incomplete outcome data. | Describe the completeness of outcome data for each main outcome, including attrition and exclusions from the analysis. State whether attrition and exclusions were reported, the numbers in each intervention group (compared with total randomised participants), reasons for attrition/exclusions where reported, and any re‐inclusions in analyses performed by the review authors. | Attrition bias due to amount, nature or handling of incomplete outcome data: High risk/Low risk/Unclear risk? |
| Selective reporting. | State how the possibility of selective outcome reporting was examined by the review authors, and what was found. | Reporting bias due to selective outcome reporting: High risk/Low risk/Unclear risk? |
| Other sources of bias. | State any important concerns about bias not addressed in the other domains in the tool. | Bias due to problems not covered elsewhere in the table: High risk/Low risk/Unclear risk? |

